3.9
TASSFUN : Target-specific scoring
functions (project
manager: RW)
People:
Stefan Henrich,
Rebecca Wade
Collaboration
Partners: Niklas
Blomberg (AstraZeneca
RD,
Sponsor: AstraZeneca
Thromboembolic diseases are common in humans. They are often affected by enzymes of the blood coagulation cascade, so the specific inhibition of these enzymes is a major goal in drug design. In this project, Comparative Binding Energy (COMBINE) analysis is being developed for generating target-specific scoring functions (TASSFUN) for blood coagulation cascade enzymes, which will be applied for virtual ligand screening and for predicting the bioactivity of new inhibitors.
COMBINE analysis relies on a training set of structures of protein-inhibitor-complexes with accompanying bioactivity values, such as inhibitor constants. For these complexes, the electrostatic and van der Waals interaction energies are calculated between the inhibitor and each protein residue. In addition, electrostatic desolvation energy terms are computed by solving the Poisson-Boltzmann equation for the protein and the inhibitor in each complex. The decomposed interaction energies and the electrostatic desolvation energy terms are correlated by Partial Least Squares (PLS) to bioactivity or binding free energy values. Subsequently, the derived target-specific scoring function is used to predict the bioactivity of inhibitors for which no experimental values are available.
On the methodological side, we have worked on streamlining the structural modeling of complexes and energetic calculations necessary for COMBINE analysis so that these steps are automated for large datasets. We have also addressed difficult issues for the prediction of the structures of protein-ligand complexes posed by the enzymes studied such as the presence of alternative protein side-chain and ligand functional group positions, water molecules in the binding site and very short hydrogen-bonds. We are assembling suitable datasets of diverse enzyme-inhibitor complexes with structural and binding affinity information and are in the process of building and testing COMBINE models for different protein targets.
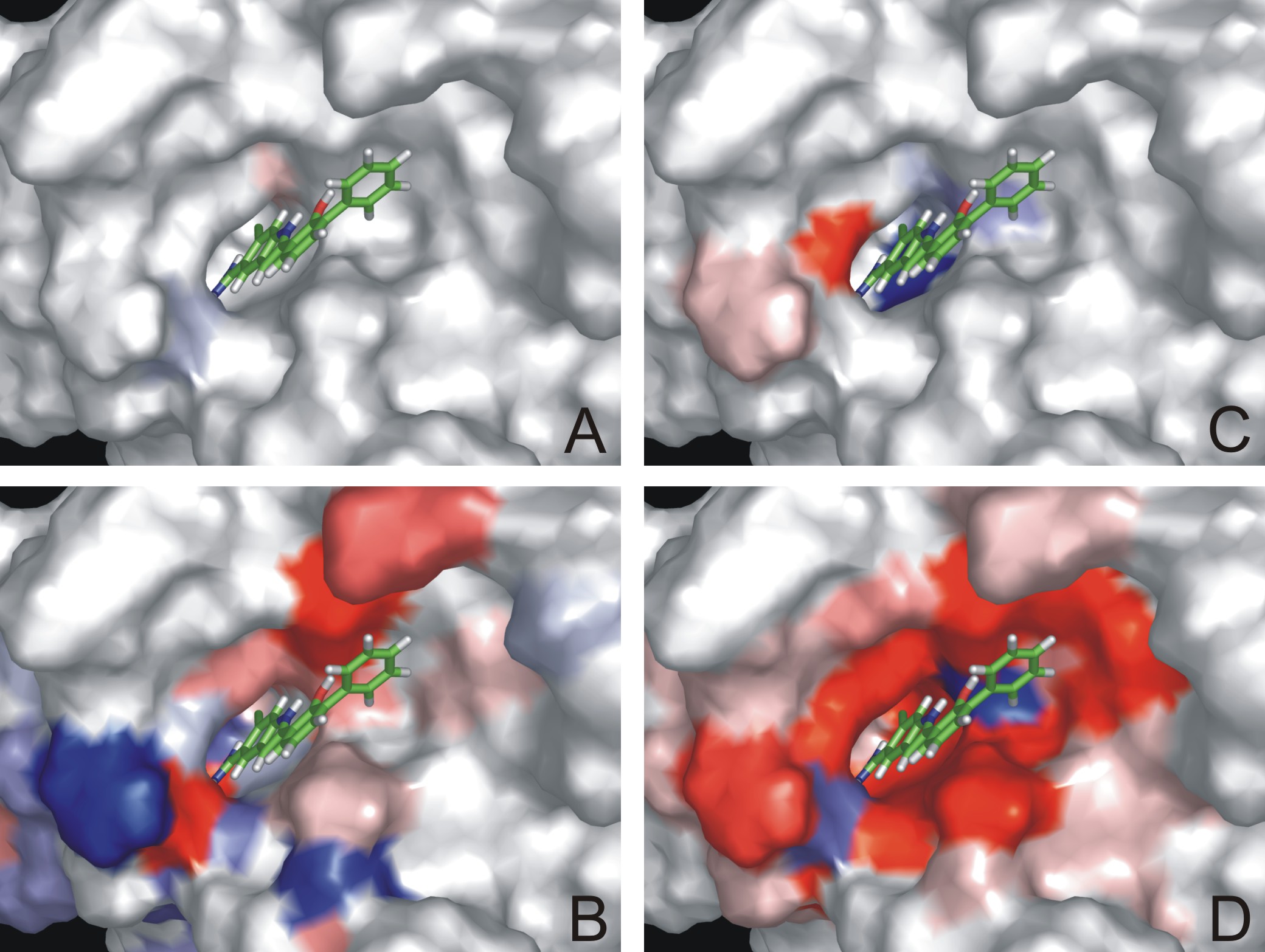
TASSFUN+F1:
Visualization of
interaction energy terms. Electrostatic (A, B) and van der Waals (C, D)
interaction values between a receptor model and one of the compounds in
the
training set were mapped onto the receptor binding site surface (in red
stabilizing and in blue destabilizing regions for complex formation).
The upper
panel (A, C) shows the unweighted
interaction energy
terms and the lower panel (B, D) the corresponding ones, weighted by
the
COMBINE model. It can be seen that not all of the interaction energies
have the
same impact to the binding affinity.
6
Professional Activities
Darmstdter Molecular Modelling Worksh