Ligands of Howard et al, JMC 2006, 49, 1346
For the ligands just IC50 values were given. These
were not used in the training sets, but the ligands could be used as an external test set.
local_start.sh -sdf
/home/henricsn/combine2go/sdf/all_071106_3D.sdf \
-inrec
../model3_180506/ -inlig ./ -model thrombin-model3.min.pdb \
-ligbase
.ligand.pdb |tee local.log
calc_inter_model.sh -ki thrombin -x -indir ./ -outdir ./ -sdf \
/home/henricsn/combine2go/sdf/all_071106_3D.sdf
-res2id residue2list|tee calc.log
Data file :
/home/henricsn/combine2go/data/thrombin/model3_180506/anal2/output-A12_1-A09-A84-A88-A89-A90.dat
no
variable selection, LV4
External
PLS predictions for
/home/henricsn/combine2go/data/thrombin/ligands_Howard/ligands_Howard.dat
Results
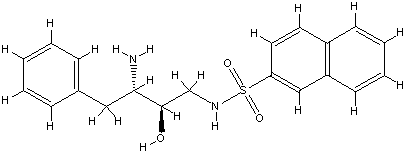
For the recently published crystal structures of thrombin, which were in complex with a series of ligands binding exclusively to the S1 and/or S2/S3/S4 pockets, IC50 but no Ki values were given. Because of the missing Ki values they were not used as a training set, but under the assumption of similar order of magnitude for IC50 and Ki values the ligands were suitable as an external test set to evaluate the COMBINE model. The predicted ∆G values for the X‑ray and docked conformation of the ligands B29/2c8w, B28/2c8x and id403 (no X-ray structure available) were similar to the experimental ones demonstrating a good quality of the COMBINE model of thrombin. But for ligands, which weakly bound in the experiment either only to the S1 pocket (B26/2c8z, B24/2c90) or left the S1 pocket vacant (B25/2c8y, B27/2c93) (Howard, et al., 2006), could not be predicted correctly as weak binders.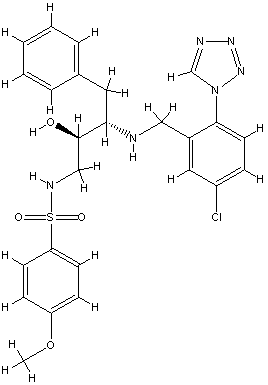
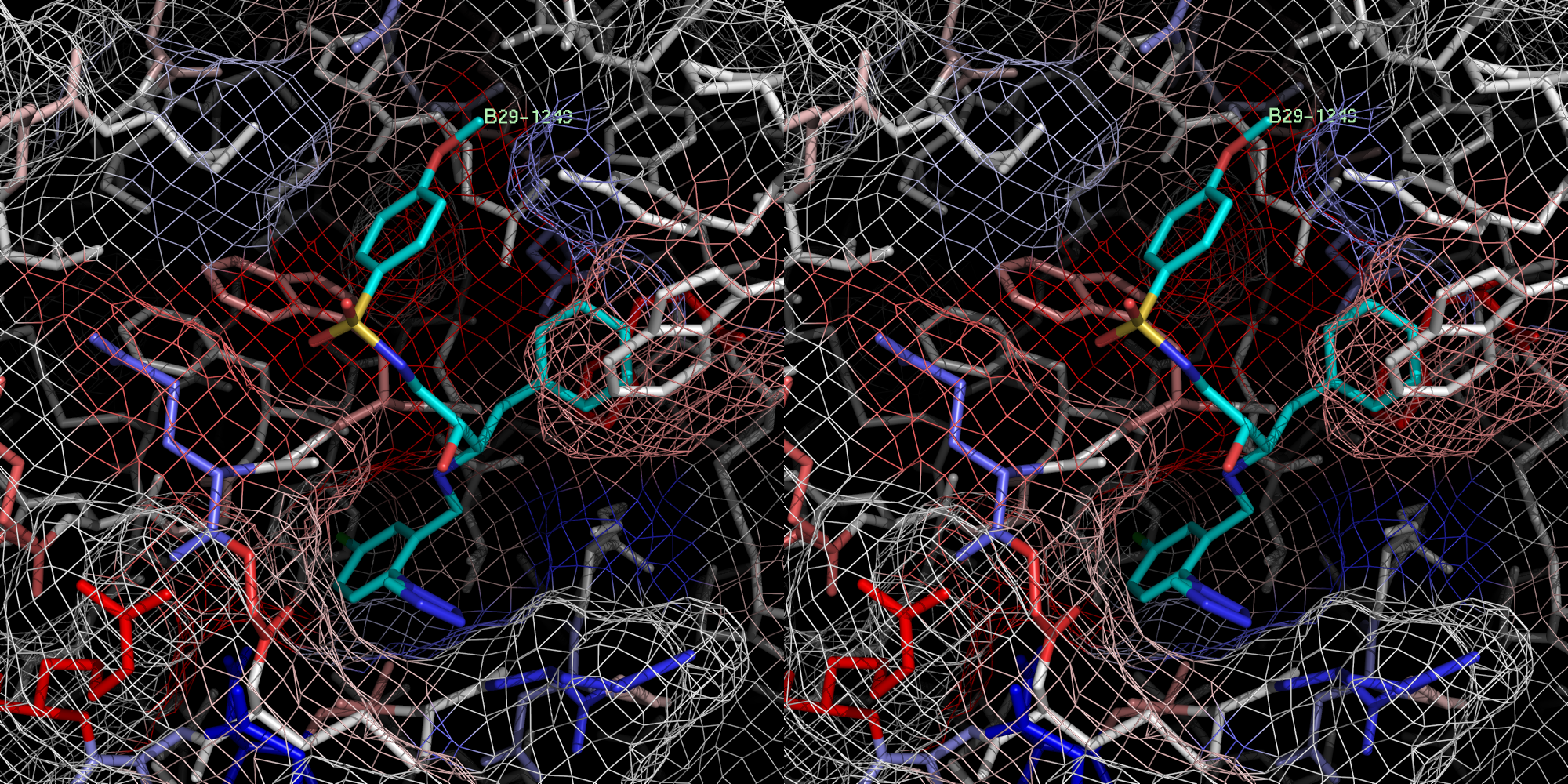
xray
1 B29.0 id 404 2c8w
Exper: +8.889e+05 Pred:( 1 ) -11.95 ( 2 ) -14.08 ( 3 ) -13.98 ( 4 ) -14.19 ( 5 ) -14.45
pred Ki = 3.96123e-05 = 0.00004 uM
exper IC50 = 0.0037 uM
exper delta G in kcal/mol= -11.5021
docking
best dG bind elec
69 id404.thr.7.9
Exper:
+8.889e+05 Pred:( 1 ) -8.795
( 2 ) -11.24 ( 3 )
-11.66 ( 4 ) -12.28 ( 5 ) -12.33
wrong
docking pose
61 id404.thr.7.1
Exper:
+8.889e+05 Pred:( 1 ) -8
( 2 ) -9.639 ( 3 )
-9.695 ( 4 ) -10.46 ( 5 ) -10.64
better
docking pose
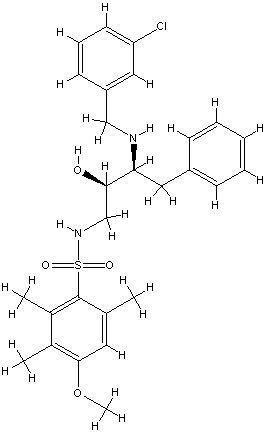
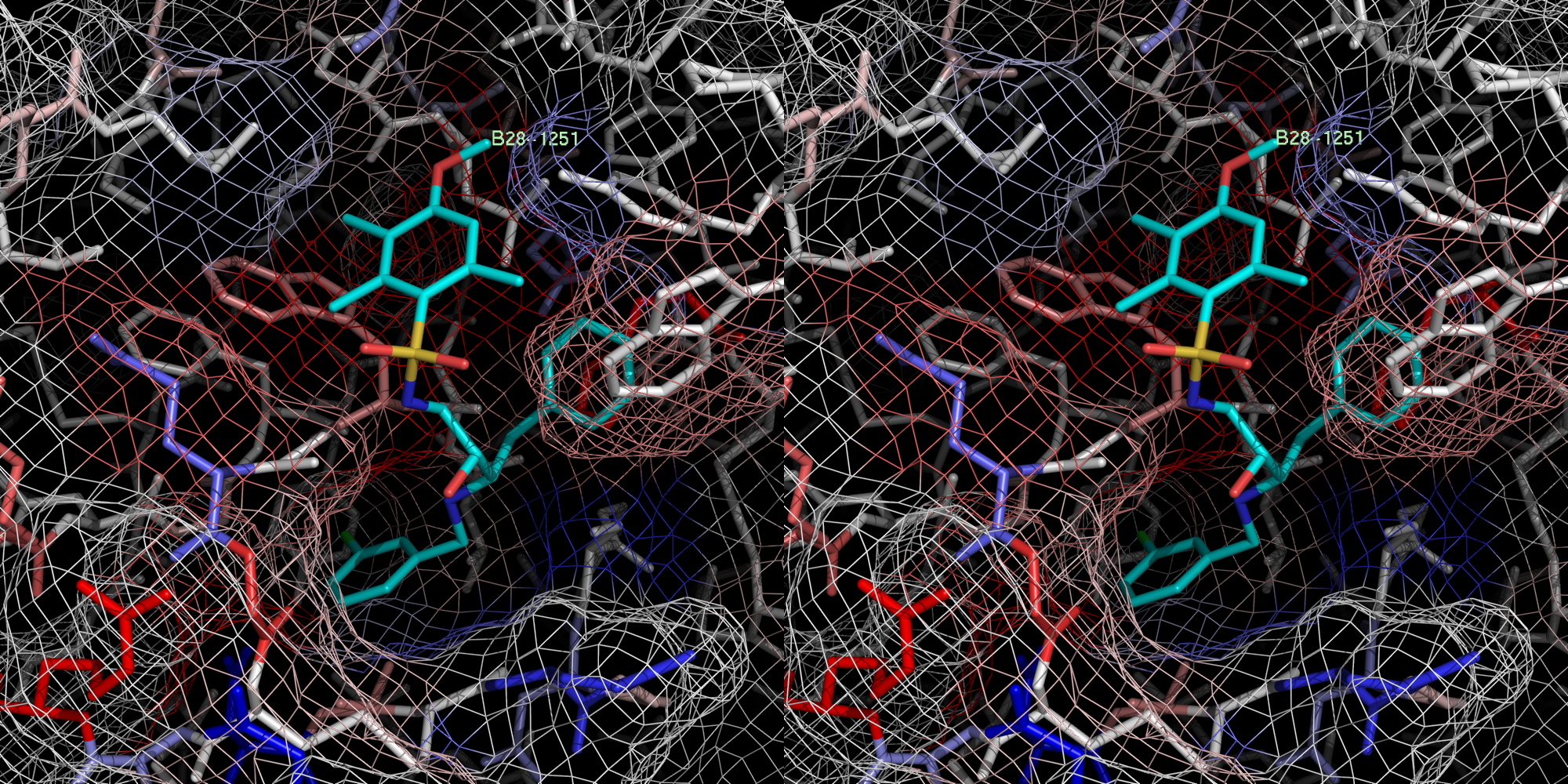
xray
2 B28.0
id 402
2c8x
Exper: +8.889e+05 Pred:( 1 ) -11.7 ( 2 ) -13.76 ( 3 ) -13.49 ( 4 ) -13.73 ( 5 ) -14.03
pred Ki = 8.61072e-05=0.00009 uM
exper IC50 = 0.22 uM
exper delta G in kcal/mol= -9.08186
docking
best dG bind elec
46 id402.thr.5.6
Exper:
+8.889e+05 Pred:( 1 ) -9.396
( 2 ) -11.68 ( 3 )
-12.37 ( 4 ) -12.62 ( 5 ) -12.37
wrong
docking pose
43 id402.thr.5.3
Exper:
+8.889e+05 Pred:( 1 ) -8.858
( 2 ) -10.95 ( 3 )
-10.69 ( 4 ) -10.29 ( 5 ) -10.07
better
docking pose
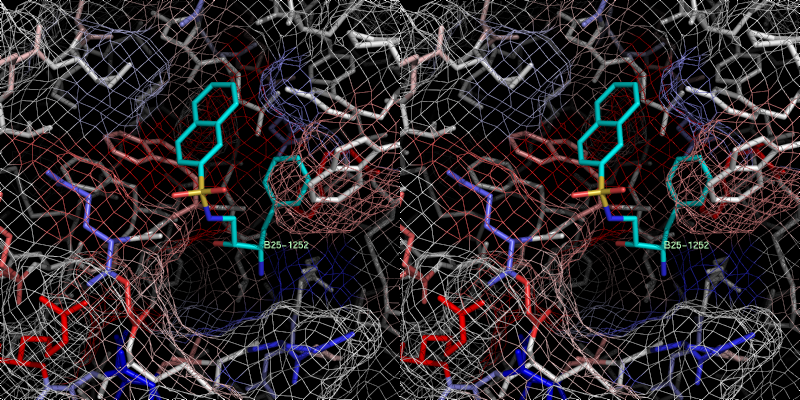
xray
3 B25.0 id 399
2c8y
Exper:
+8.889e+05 Pred:( 1 ) -9.896
( 2 ) -11.75 ( 3 )
-11.41 ( 4 ) -11.52 ( 5 ) -11.41
pred Ki = 0.00359012
exper IC50 = 100 uM
exper delta G in kcal/mol= -5.45655
docking
best dG bind elec
19 id399.thr.2.9
Exper:
+8.889e+05 Pred:( 1 ) -8.417
( 2 ) -10.25 ( 3 )
-10.26 ( 4 ) -11.01 ( 5 ) -11.04
wrong
docking pose
The bad prediction is propably based on the vacant S1 pocket.
docking
pose in high energetic open conformation but in Xray
a hydrophobic collapse was observed
fragment
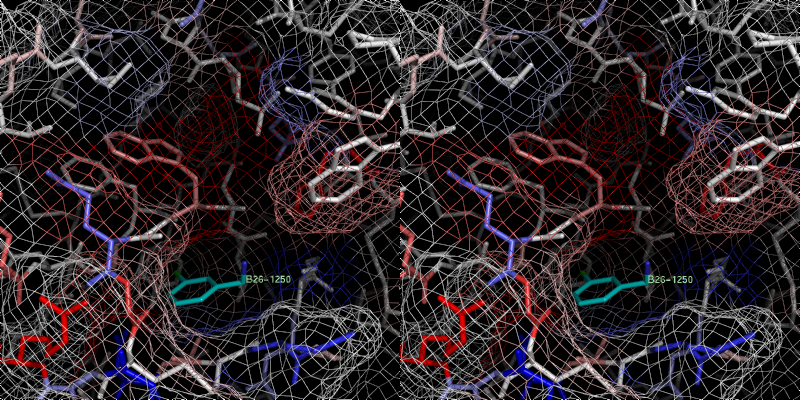
xray
4 B26.0 id 400 2c8z
Exper: +8.889e+05 Pred:( 1 ) -7.298 ( 2 ) -8.516 ( 3 ) -7.716 ( 4 ) -8.242 ( 5 ) -8.203
pred Ki =
0.908012
exper IC50 >
1000 uM
exper delta G in
kcal/mol < -4.09
docking
best dG bind elec
26 id400.thr.3.6
Exper:
+8.889e+05 Pred:( 1 ) -6.372
( 2 ) -6.114 ( 3 )
-6.124 ( 4 ) -6.001 ( 5 ) -5.823
wrong
docking pose
fragment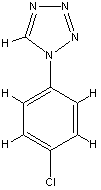
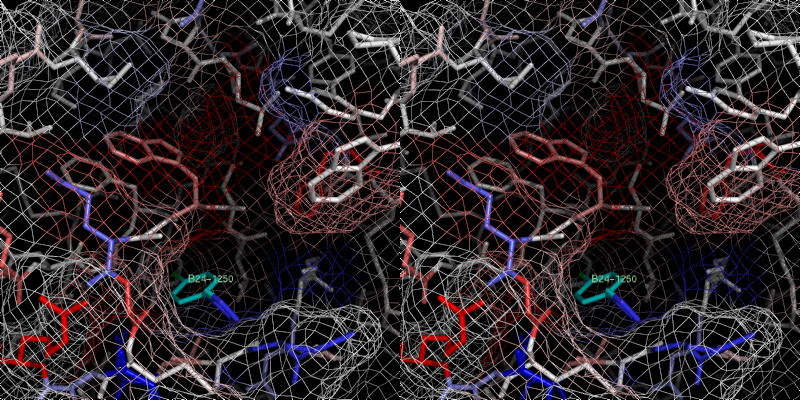
xray
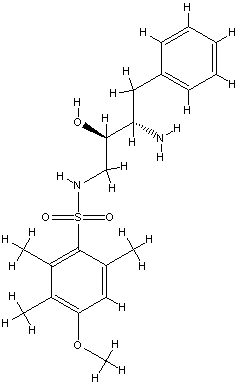
5 B24.0 id 398
2c90
Exper:
+8.889e+05 Pred:( 1 ) -7.826
( 2 ) -10.19 ( 3 )
-9.976 ( 4 ) -10.1 ( 5 ) -9.992
pred Ki = 0.0394511
exper IC50 = 330 uM
exper delta G in
kcal/mol= -4.74923
docking
best dG bind elec
4 id398.thr.1.4
Exper:
+8.889e+05 Pred:( 1 ) -7.944
( 2 ) -9.953 ( 3 )
-9.815 ( 4 ) -9.727 ( 5 ) -9.565
wrong
docking pose
The prediction for this small fragment is bad, but
e. g. benzamidine with similar size were not used in the training set,
too, becaue of beeing an outlier or an inconsistancy in the Ki values.
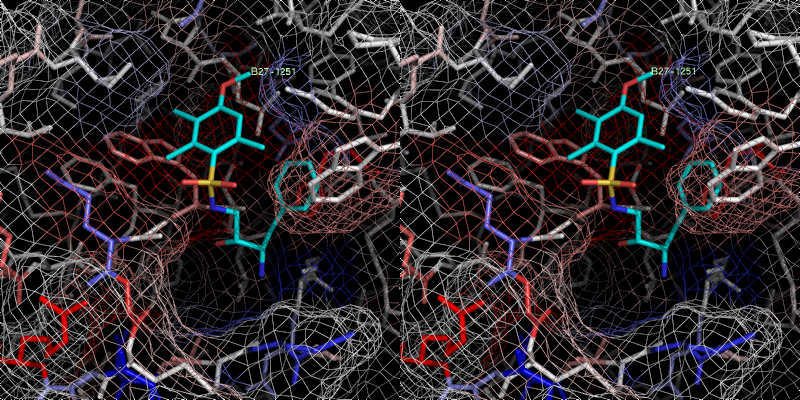
xray
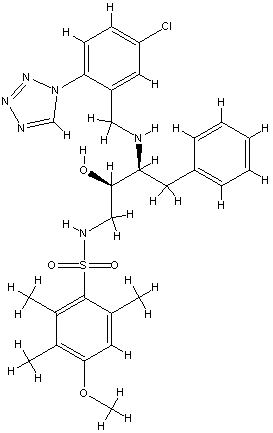
6 B27.0 id 401 2c93
Exper: +8.889e+05 Pred:( 1 ) -10.12 ( 2 ) -11.9 ( 3 ) -11.4 ( 4 ) -11.32 ( 5 ) -11.24
pred Ki =
0.00503178
exper IC50 = 12
uM
exper delta G in
kcal/mol= -6.71268
docking
best dG bind elec
38 id401.thr.4.8
Exper:
+8.889e+05 Pred:( 1 ) -8.167
( 2 ) -10.39 ( 3 ) -10.5 ( 4 ) -11.14
( 5 ) -11.34
wrong
docking pose
Bad prediction due to a vacant S1 pocket (similar to B25).
no xray available
id
403
exper IC50 = 0.0014 uM
exper delta G in
kcal/mol= -12.0779
docking
51 id403.thr.6.1
Exper:
+8.889e+05 Pred:( 1 ) -8.238
( 2 ) -10.11 ( 3 )
-10.27 ( 4 ) -10.42 ( 5 ) -10.31
wrong
docking pose
53 id403.thr.6.3
Exper: +8.889e+05 Pred:( 1 ) -9.193 ( 2 ) -11.22 ( 3 ) -11.18 ( 4 ) -11.52 ( 5 ) -11.65
best docking pose