| ligand |
PDB |
deltaG exp |
deltaG exp | pred deltaG, w/o variable selection, LV4 |
||
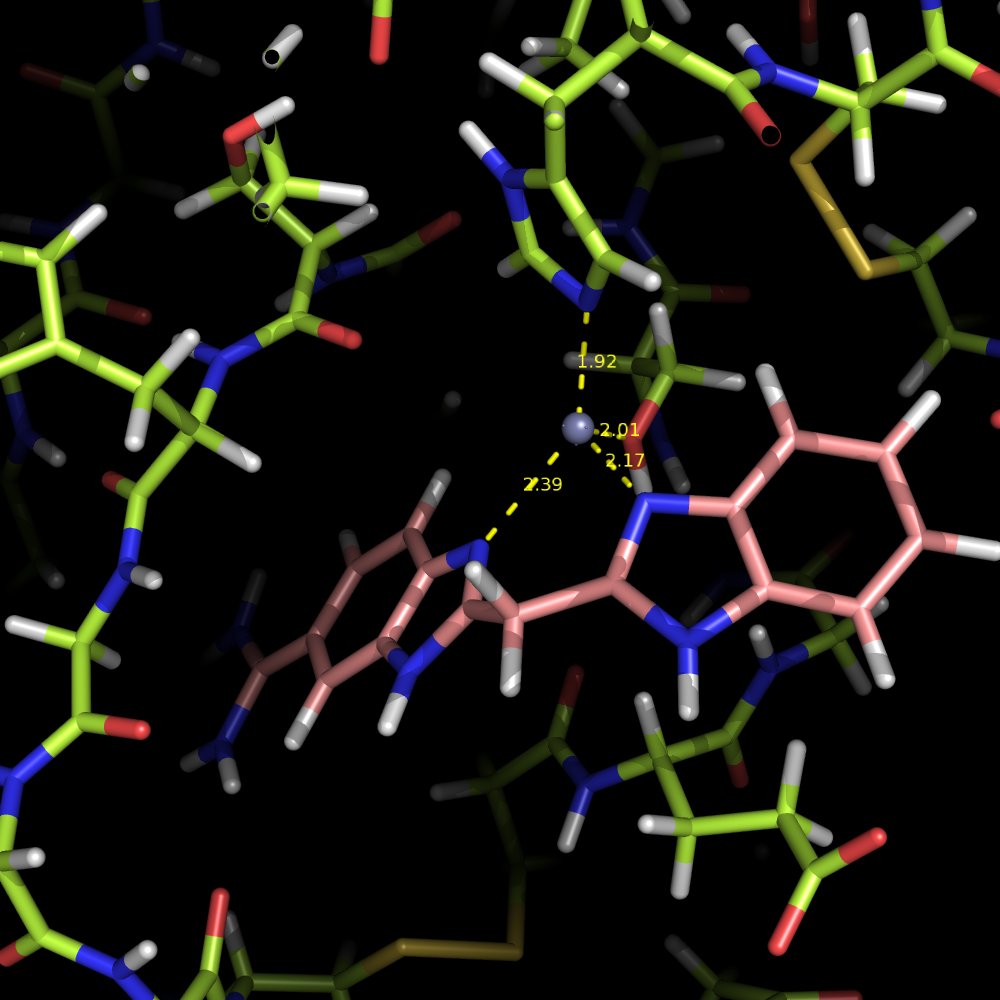
| EDTA |
Zn2+ |
X-ray w/o Zn |
X-ray with fixed Zn | docking best deltaG desolv (UHBD) ranked |
||
| A49 |
1c1u |
-7.7 |
-11.2 |
-7.3 |
-6.3 |
-8.7 |
| A58 |
1c1v |
-7.4 |
-10.4 |
-5.6 |
-4.8 |
-7.0 |
| A59 |
1c1w |
-4.1 |
-9.7 |
-5.9 |
n.d. |
-6.6 |