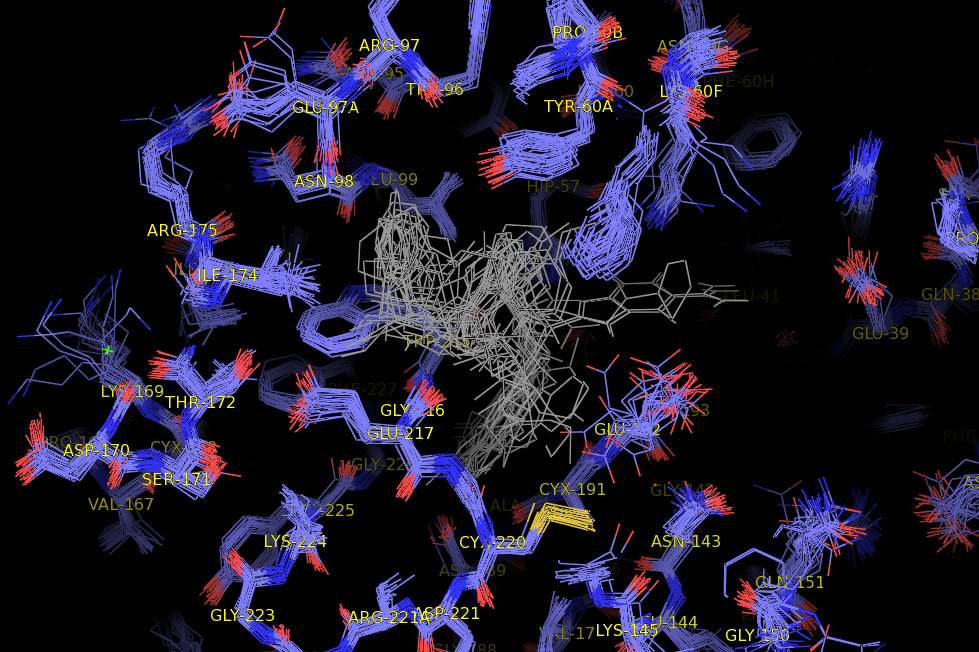
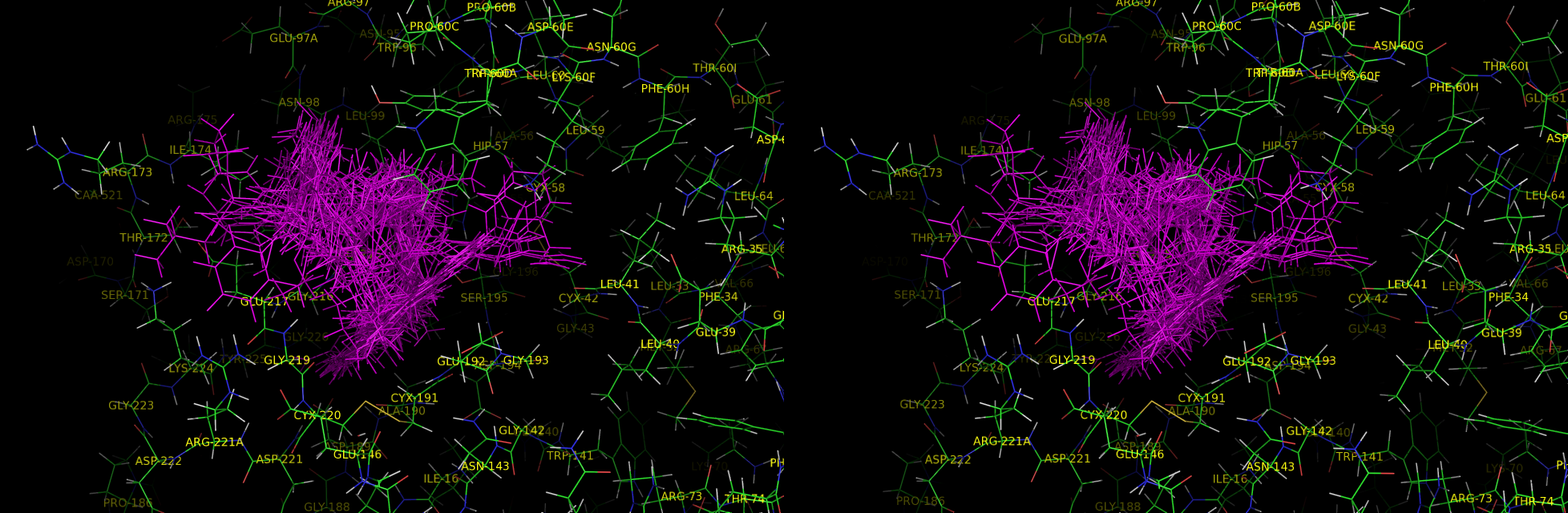
procedure for preparing a thrombin model
1. Superposition 2. Building a receptor model xleap.sh -pdb thrombin-model -n 000
-prot thrombin-model-temp.pdb -lig model1.ligand -m
/home/henricsn/combine2go/parameter/protmin.thrombin-model.template -w
-noac -nopc -notemp -bres |tee xleap.log thrombin-model.min.pdb thrombin-model.minbres.pdb 3. Ki values all_160206_3D.sdf 4. Preparation of ligands 1a4w.ligand.prot.pdb
1c1v.ligand.prot.pdb 1dwd.ligand.prot.pdb
1o5g.ligand.prot.pdb 1ae8.ligand.prot.pdb
1c1w.ligand.prot.pdb 1fpc.ligand.prot.pdb
1qbv.ligand.prot.pdb 1afe.ligand.prot.pdb
1c4u.ligand.prot.pdb 1ghv.ligand.prot.pdb
1ta2.ligand.prot.pdb 1aix.ligand.prot.pdb
1c4v.ligand.prot.pdb 1ghw.ligand.prot.pdb
1ta6.ligand.prot.pdb 1bcu.ligand.prot.pdb
1c4y.ligand.prot.pdb 1ghy.ligand.prot.pdb
1tom.ligand.prot.pdb 1bhx.ligand.prot.pdb
1c5n.ligand.prot.pdb 1gj4.ligand.prot.pdb
7kme.ligand.prot.pdb 1bmm.ligand.prot.pdb
1d6w.ligand.prot.pdb 1k21.ligand.prot.pdb
8kme.ligand.prot.pdb 1bmn.ligand.prot.pdb
1d9i.ligand.prot.pdb 1k22.ligand.prot.pdb 1c1u.ligand.prot.pdb
1dwc.ligand.prot.pdb 1o2g.ligand.prot.pdb pdbconv-ligand
1a4w.ligand.prot.pdb 1a4w.ligand.pdb
5. Minimization local_start.sh
-sdf /home/henricsn/combine2go/sdf/all_160206_3D.sdf
-inrec ./ -inlig /home/henricsn/combine2go/data/thrombin/pdb/ligand/
-model thrombin-model.min.pdb -ligbase .ligand.pdb |tee local.log not selected structures because of
coordinates Zn in active site: 6. Interaction energies 29
ligands: cd
anal/ 7. Prediction
Data file :
/home/henricsn/combine2go/data/thrombin/model1_200206/golpe/output.dat
4
4.8495 73.2061
4
5.3508e+03 6.3855e+02
8.3797
4
2.3666 66.3532
1.1146 0.8155
4
2.2379
- 0.2562
3
7.6649 59.4839
1.2325 0.7744
3
1.8103
- 0.5133 after removing potential
outlier: A91 Data file :
/home/henricsn/combine2go/data/thrombin/model1_200206/golpe/output-A91.dat
4
4.1313 73.8963
4
5.4931e+03 5.8741e+02
9.3513
4
3.1321 67.6472
1.0038 0.8534
4
1.9244
- 0.4611
3
9.6057 65.1034
3
3.6891e+02 3.2678e+02
1.1289
3
6.4920 59.5700
1.0813 0.8299
3
1.5735
- 0.6397 Privacy Imprint