receptor: urokinase model5-cut.pdb
ligands: X-ray
conformations of urokinase complexes
minimization: medium
constraints
UHBD input: desolv1_35.inp, desolv2_35.inp
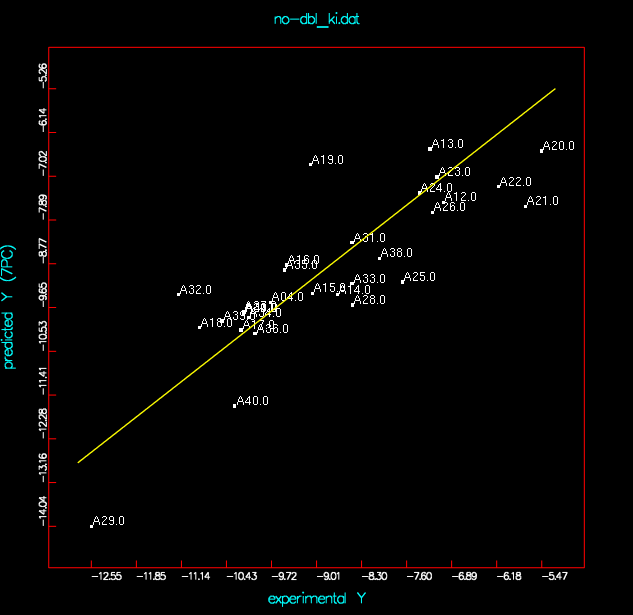
| no-dbl-Ki |
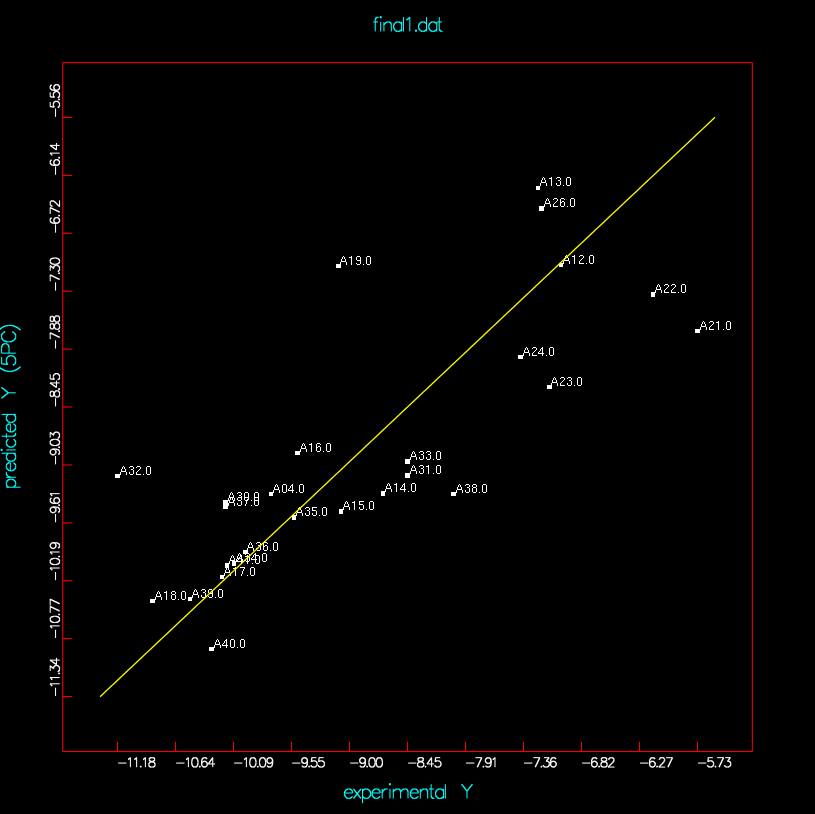
no-dbl_ki-A20-A29 | final1 |
Katz |
Strzebecher | Wendt | set1 |
set2 |
set3 |
set4 |
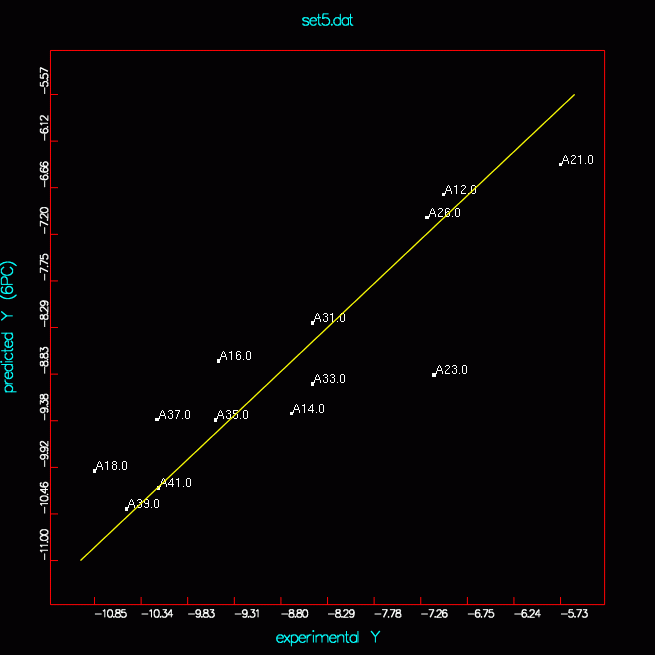
set5 |
| 30 |
28 |
26 |
12 |
6 |
9 |
17 |
17 |
18 |
13 |
13 |
| A04 A12 A13 A14 A15 A16 A17 A18 A19 A21 A20 A22 A23 A24 A25 A26 A28 A29 A30 A31 A32 A33 A34 A35 A36 A37 A38 A39 A40 A41 |
A04 A12 A13 A14 A15 A16 A17 A18 A19 A21 A22 A23 A24 A25 A26 A28 A30 A31 A32 A33 A34 A35 A36 A37 A38 A39 A40 A41 |
A04 A12 A13 A14 A15 A16 A17 A18 A19 A21 A22 A23 A24 A26 A30 A31 A32 A33 A34 A35 A36 A37 A38 A39 A40 A41 |
A04 A12 A13 A14 A15 A16 A17 A18 A19 A21 A23 A24 |
A25 A26 A28 A39 A40 A41 |
A30 A31 A32 A33 A34 A35 A36 A37 A38 |
A12 A13 A15 A16 A18 A19 A22 A23 A26 A30 A32 A33 A35 A36 A38 A39 A41 |
A04 A13 A14 A16 A17 A19 A21 A23 A24 A30 A31 A33 A34 A36 A37 A39 A40 |
A04 A12 A14 A15 A17 A18 A21 A22 A24 A26 A31 A32 A34 A35 A37 A38 A40 A41 |
A04 A13 A15 A17 A19 A22 A24 A30 A32 A34 A36 A38 A40 |
A12 A14 A16 A18 A21 A23 A26 A31 A33 A35 A37 A39 A41 |
| file | # |
LV |
XAccum |
R |
SDEC | r2 |
SDEP |
q2 |
selection |
LV |
XAccum |
R |
SDEC |
r2 |
SDEP | q2 |
| no-dbl_ki | 30 |
1 2 3 4 5 6 7 8 9 10 |
44.5509 62.5665 73.1424 79.2187 83.0818 86.0039 88.4328 90.9105 92.5376 93.6349 |
0.6461 0.8995 1.0990 0.9792 1.5743 2.0828 1.9133 1.0747 1.0068 1.1036 |
1.4039 1.1246 0.9264 0.7632 0.6897 0.5901 0.5300 0.4972 0.4379 0.4110 |
0.2998 0.5508 0.6951 0.7931 0.8310 0.8763 0.9002 0.9122 0.9319 0.9400 |
1.6044 1.5527 1.5970 1.5976 1.5521 1.4673 1.4666 1.4454 1.5122 1.6160 |
0.0855 0.1436 0.0940 0.0933 0.1443 0.2352 0.2360 0.2578 0.1877 0.0723 |
D8F8 no-dbl_ki.ps |
1 2 3 4 5 6 7 8 9 10 |
33.9779 53.8904 71.2340 80.9194 85.1923 88.2825 90.3499 92.1007 93.5181 94.7112 |
0.8397 1.2544 0.8852 0.7887 6.1648 1.4612 1.9452 1.0862 1.4179 1.1035 |
1.2877 1.0862 0.7906 0.6707 0.5952 0.5863 0.5621 0.5158 0.4496 0.4089 |
0.4110 0.5809 0.7780 0.8402 0.8742 0.8779 0.8878 0.9055 0.9282 0.9406 |
1.5745 1.4683 1.2686 1.1368 1.0382 0.9756 0.9493 1.1016 1.2994 1.4816 |
0.1193 0.2341 0.4283 0.5409 0.6171 0.6619 0.6798 0.5689 0.4002 0.2202 |
| no-dbl_ki-A20-A29 |
28 |
0.0172 |
||||||||||||||
| final1 (test5-A25-28.dat) |
26 |
1 2 3 4 5 6 7 8 9 10 |
48.2587 68.1613 77.3882 81.8637 84.9804 87.7089 89.9040 92.2830 93.4791 94.6019 |
0.6263 0.8132 8.9280 1.4642 1.4189 1.4426 1.2385 1.2359 1.5429 1.1159 |
1.2628 0.9951 0.8629 0.7404 0.6413 0.5611 0.5109 0.4375 0.4014 0.3364 |
0.2805 0.5532 0.6640 0.7527 0.8144 0.8579 0.8822 0.9136 0.9273 0.9489 |
1.4295 1.3213 1.2529 1.1931 1.3708 1.5552 1.8249 2.1079 2.2369 2.5542 |
0.0779 0.2123 0.2917 0.3578 0.1521 -0.0913 -0.5027 -1.0047 -1.2576 -1.9436 |
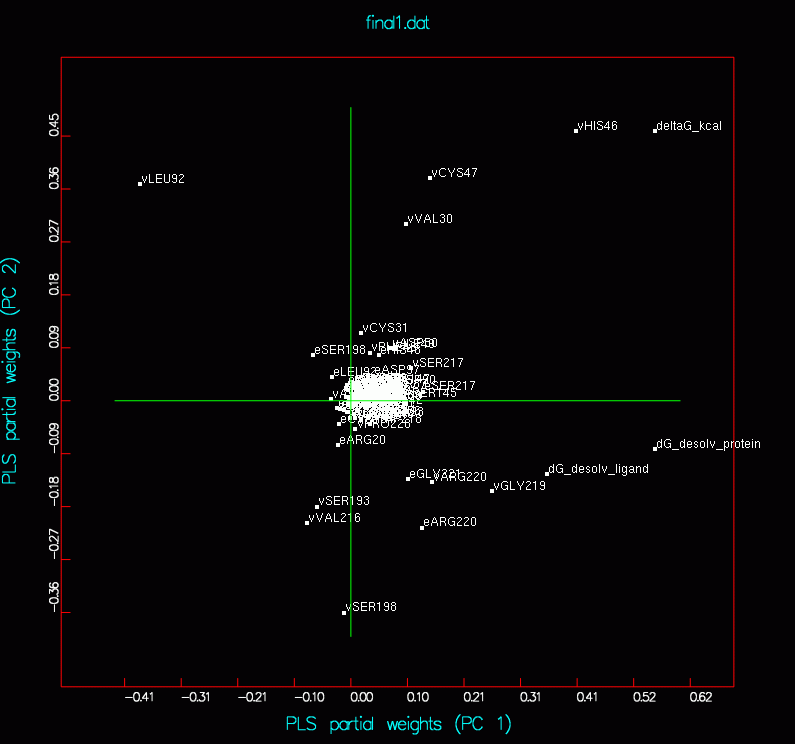
D4F4 final1_d4f4_ pred5.png final1_d4f4_ partial_weight.png |
1 2 3 4 5 6 7 8 9 10 |
53.9784 75.5756 86.4838 90.2410 91.8186 93.3859 94.5115 95.3955 96.2554 96.8500 |
0.6124 0.6832 1.4816 0.9329 5.4205 1.9384 25.2159 1.2684 0.9606 1.3176 |
1.2422 0.9707 0.8430 0.7099 0.6200 0.5415 0.4607 0.4049 0.3541 0.3268 |
0.3037 0.5749 0.6793 0.7726 0.8266 0.8677 0.9042 0.9260 0.9434 0.9518 |
1.3930 1.2094 1.1086 0.9567 0.9183 0.9549 1.0052 1.0801 1.1080 1.1382 |
0.1245 0.3400 0.4454 0.5870 0.6195 0.5886 0.5441 0.4737 0.4461 0.4155 |
| final1-buw |
3 |
0.2484 |
with BUW no better results than
without |
|||||||||||||
| Katz | 12 |
0.0830 |
||||||||||||||
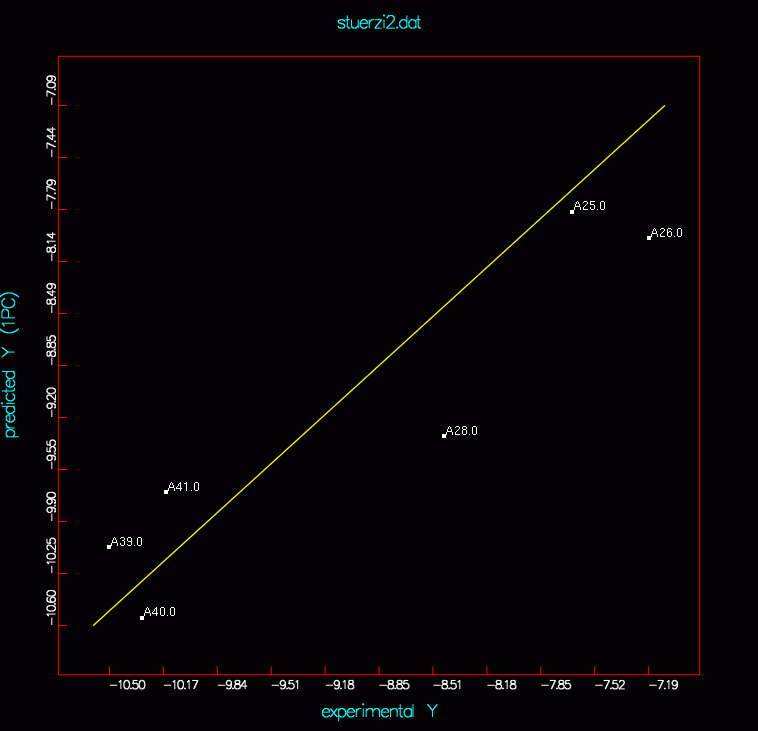
| Strzebecher | 6 |
1 2 3 4 |
49.1131 73.7768 90.2656 95.4428 |
0.7401 34.8902 0.2648 0.5645 |
0.2856 0.1466 0.0749 0.0013 |
0.9538 0.9878 0.9968 1.0000 |
0.5579 0.5459 0.5720 0.5419 |
0.8238 0.8313 0.8148 0.8338 |
stuerzi2_pred1.png |
|||||||
| Wendt | 9 |
- |
||||||||||||||
| set1 |
17 |
3 |
0.1169 |
|||||||||||||
| set2 |
17 |
4 |
0.7910 |
1.2028 |
0.2239 |
|||||||||||
| set3 |
18 |
4 |
80.9451 |
0.5741 |
0.8703 |
1.0991 |
0.5249 |
|||||||||
| set4 |
13 |
- |
||||||||||||||
| set5 |
13 |
1 2 3 4 5 |
50.8217 69.6755 77.4136 83.4992 88.3866 |
0.5609 0.8637 10.3950 17.8731 211.3251 |
1.1740 0.7278 0.5692 0.4058 0.2601 |
0.4022 0.7702 0.8595 0.9286 0.9707 |
1.5062 1.2734 1.2187 1.1978 1.2487 |
0.0161 0.2967 0.3558 0.3778 0.3237 |
prediction for
set4 D4F4 set5_d4f4_pred.png no better prediction for set4 |
1 2 3 4 5 6 |
67.6303 77.7384 84.6323 89.7860 92.5998 94.4095 |
0.3589 0.8189 1.0108 3.9976 1.9021 1.0732 |
1.2235 0.5357 0.3868 0.2680 0.1773 0.1095 |
0.3507 0.8755 0.9351 0.9689 0.9864 0.9948 |
1.4715 0.9963 0.8274 0.8142 0.7274 0.6962 |
0.0609 0.5694 0.7031 0.7125 0.7705 0.7898 |