|
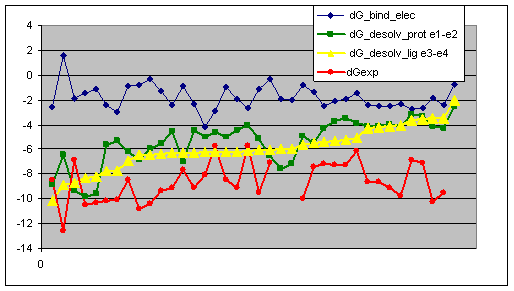
Analyzing of the desolvation energy
terms of UHBD calculated for minimized complexes receptor: urokinase model5-cut ligands: X-ray
conformations of urokinase complexes UHBD input: desolv1_35.inp, desolv2_35.inp The table is colored
according to clusters of ligands. |
|
kcal/mol |
|
|
|
|
|
res |
dG_bind_elec |
dG_desolv_prot
e1-e2 |
dG_desolv_lig
e3-e4 |
dGexp |
|
A28.0 |
-2.59549844 |
-8.85546875 |
-10.1611328 |
-8.445 |
|
A29.0 |
1.55834199 |
-6.4765625 |
-8.89147949 |
-12.55 |
|
A41.0 |
-1.88738438 |
-9.33984375 |
-8.70117188 |
-6.817 |
|
A39.0 |
-1.45603047 |
-9.8203125 |
-8.29345703 |
-10.5 |
|
A40.0 |
-1.10678359 |
-9.55078125 |
-8.18603516 |
-10.3 |
|
A30.0 |
-2.44189043 |
-5.609375 |
-7.75793457 |
-10.17 |
|
A34.0 |
-3.02483594 |
-5.30859375 |
-7.74707031 |
-10.09 |
|
A31.0 |
-0.90768672 |
-6.171875 |
-6.90673828 |
-8.454 |
|
A18.0 |
-0.85641709 |
-6.83984375 |
-6.45513916 |
-10.75 |
|
A37.0 |
-0.30779727 |
-5.9375 |
-6.43530273 |
-10.4 |
|
A32.0 |
-1.29965938 |
-5.52734375 |
-6.34179688 |
-9.324 |
|
A19.1 |
-2.44056006 |
-4.48046875 |
-6.29547119 |
-9.105 |
|
A25.0 |
-0.92386982 |
-7.0390625 |
-6.28936768 |
-7.662 |
|
A19.0 |
-2.38279414 |
-4.3984375 |
-6.25756836 |
-9.105 |
|
A38.0 |
-4.22392686 |
-4.99609375 |
-6.22637939 |
-8.021 |
|
A21.1 |
-2.88099756 |
-4.58984375 |
-6.22515869 |
-5.727 |
|
A33.0 |
-1.01586396 |
-5.03125 |
-6.20318604 |
-8.453 |
|
A19.4 |
-1.98355801 |
-4.421875 |
-6.19616699 |
-9.105 |
|
A21.0 |
-2.64414434 |
-4.0625 |
-6.10705566 |
-5.727 |
|
A35.0 |
-1.10913105 |
-5.16796875 |
-6.0802002 |
-9.521 |
|
A23.0 |
-0.32399893 |
-6.55859375 |
-6.07000732 |
-7.12 |
|
A42.0 |
-1.9583792 |
-7.60546875 |
-5.99835205 |
|
|
A42.1 |
-1.99076875 |
-7.20703125 |
-5.9375 |
|
|
A36.0 |
-0.85857158 |
-4.90625 |
-5.63787842 |
-9.979 |
|
A24.0 |
-1.36442637 |
-5.46484375 |
-5.45812988 |
-7.39 |
|
A26.0 |
-2.54067031 |
-4.30859375 |
-5.38183594 |
-7.193 |
|
A20.1 |
-2.14634702 |
-3.6875 |
-5.34805298 |
-7.228 |
|
A20.0 |
-1.94860278 |
-3.48828125 |
-5.27511597 |
-7.228 |
|
A22.0 |
-1.42438398 |
-3.8359375 |
-5.11547852 |
-6.147 |
|
A14.0 |
-2.42758359 |
-4.109375 |
-4.25244141 |
-8.654 |
|
A14.1 |
-2.47073496 |
-4.078125 |
-4.19104004 |
-8.654 |
|
A15.0 |
-2.53732549 |
-3.9921875 |
-4.10418701 |
-9.077 |
|
A04.0 |
-2.31456504 |
-4.1640625 |
-4.00427246 |
-9.739 |
|
A12.0 |
-2.72584236 |
-3.19140625 |
-3.62567139 |
-6.886 |
|
A13.0 |
-2.63288232 |
-3.296875 |
-3.55499268 |
-7.12 |
|
A17.0 |
-1.87034395 |
-4.12109375 |
-3.4901123 |
-10.2 |
|
A16.0 |
-2.42509736 |
-4.24609375 |
-3.46160889 |
-9.488 |
|
A27.0 |
-0.73484666 |
-2.484375 |
-2.00579834 |
|