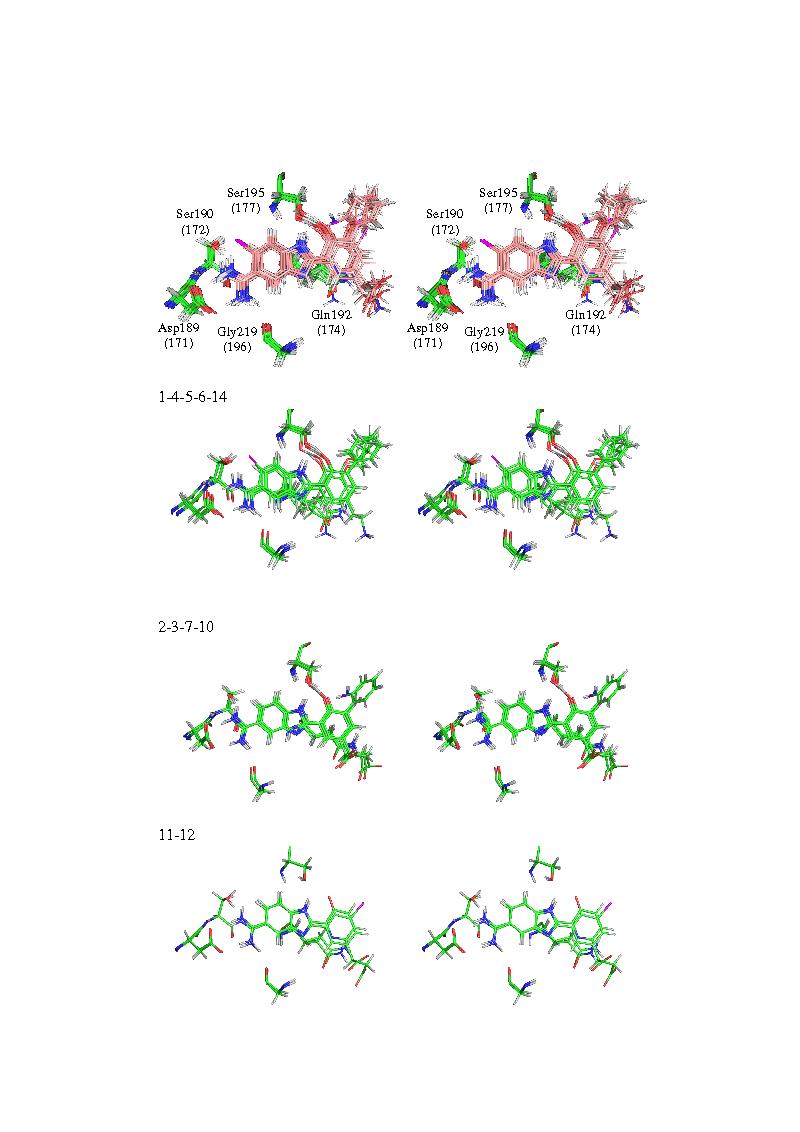
1-224 VDW
225-448 electrostatic
449 deltaG
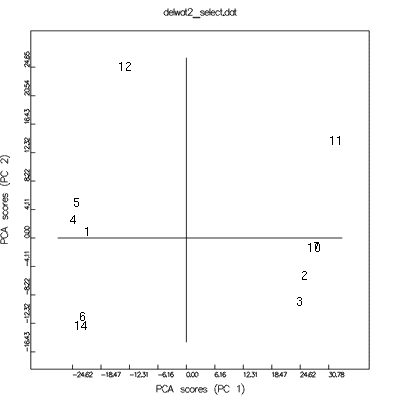
PCA-scores (x axis=1, y axis=2)
(numbers are related to #anal)
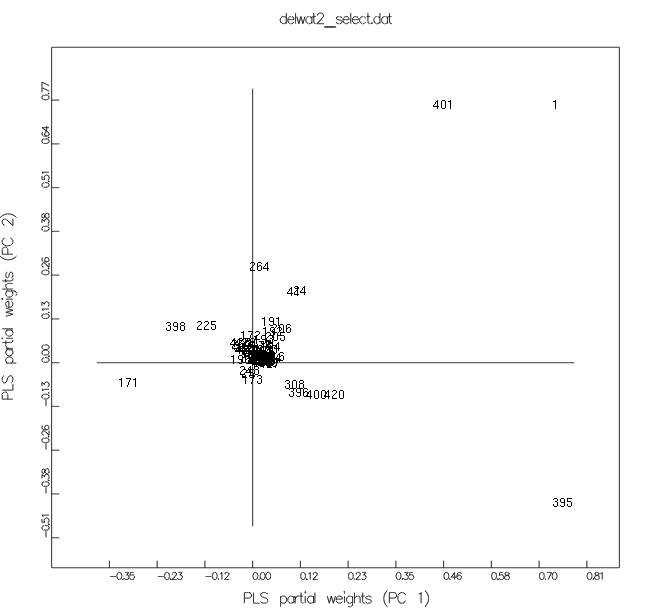
PLS partial weights
| variable |
residue |
| 171/395 |
Asp189 |
| 172/396 |
Ser190 |
| 174/398 |
Gln192 |
| 177/401 |
Ser195 |
| 196/420 |
Gly219 |
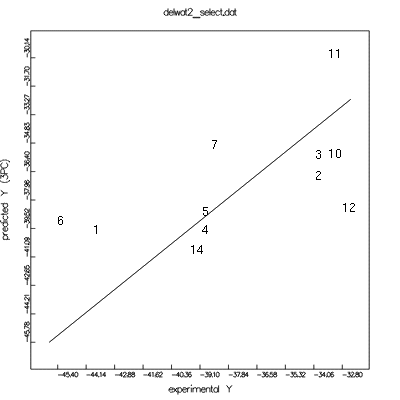
Prediction plot
(numbers are related to #anal)
Stereo figures of selected residues (pdf)