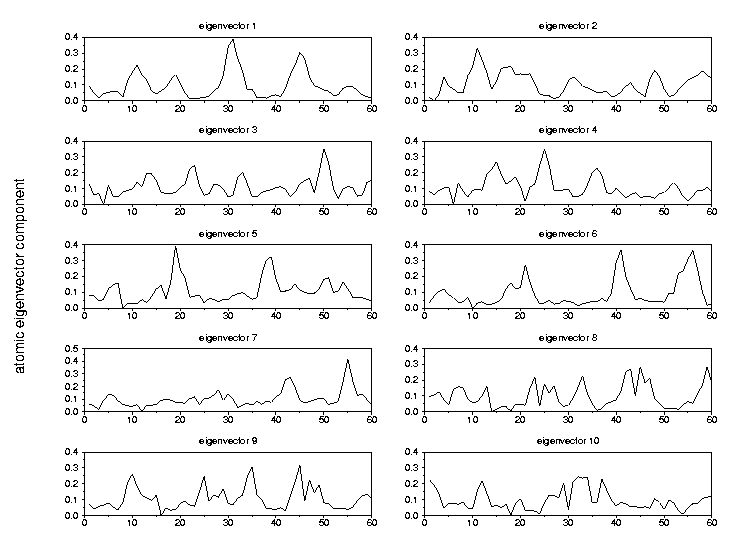
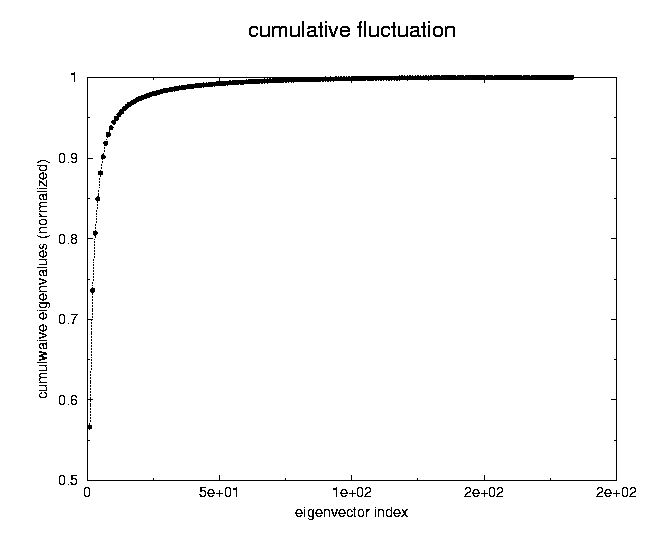
|
|
|
resolution |
|
|
|
|
|
|
 |
|
|
|
|
|
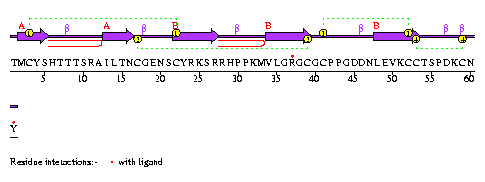 |
|
|
|
|
resolution |
|
|
|
|
|
|
 |
|
|
|
|
|
 |
|
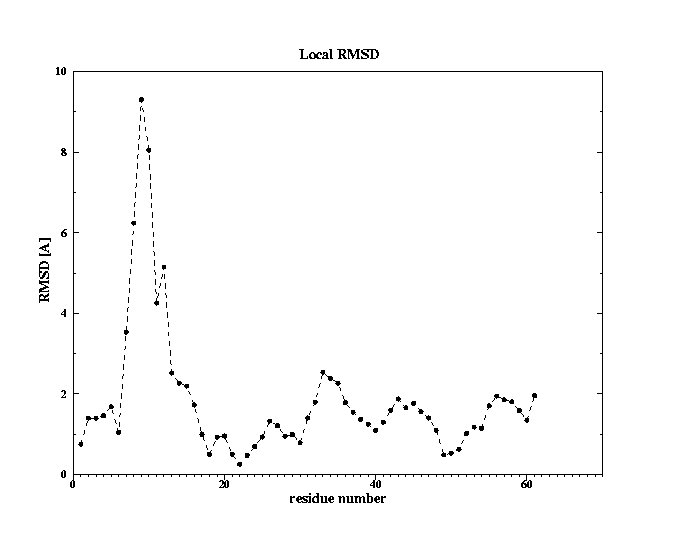

Fig. 2 Conformational change observed in loop1; fasciculin I (1fas) -red; fasciculin II (1fsc) -blue
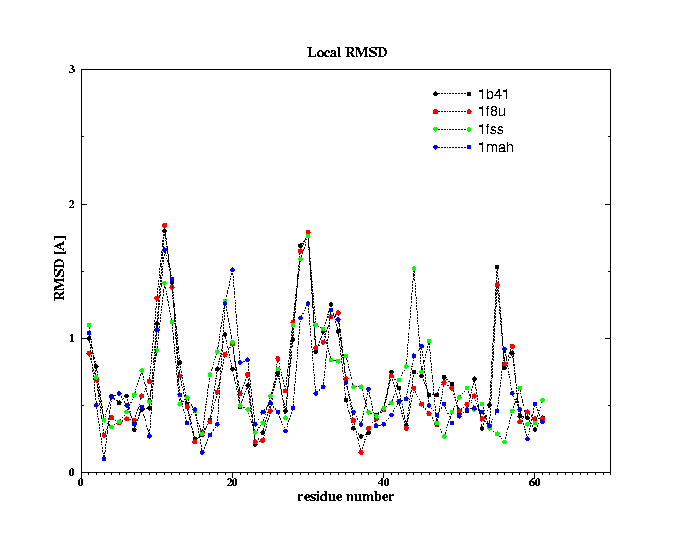
3. RMSD between the unbound and bound states of fasciculin II
Global RMSD after fitting backbone atoms
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
number of configurations generated with concoord=> 500Fig. 4 The global fluctuations in atomic motions along the configurations generated by concoord are, as expected larger4.1 Fasciculin I (pdb 1fas)
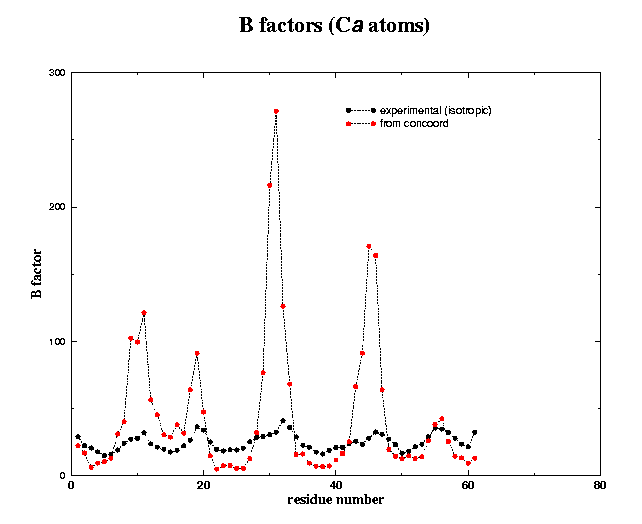
4.1.1 global motions (B factors)
4.1.2 Global motions (RMSD between the structures generated by concoord and the crystal structure of fasciculin I, pdb 1fas)
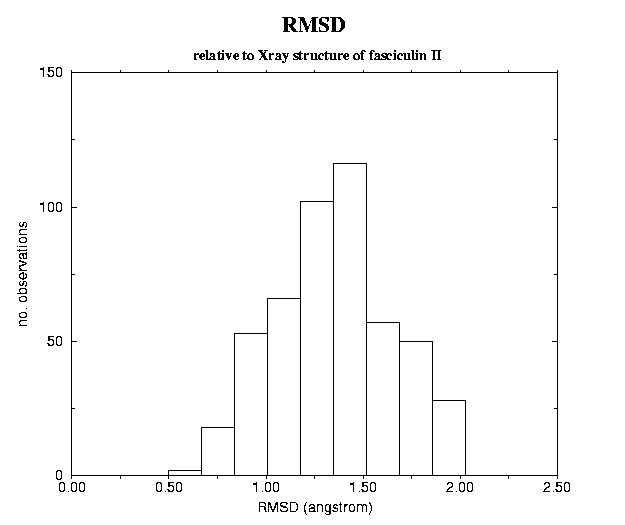
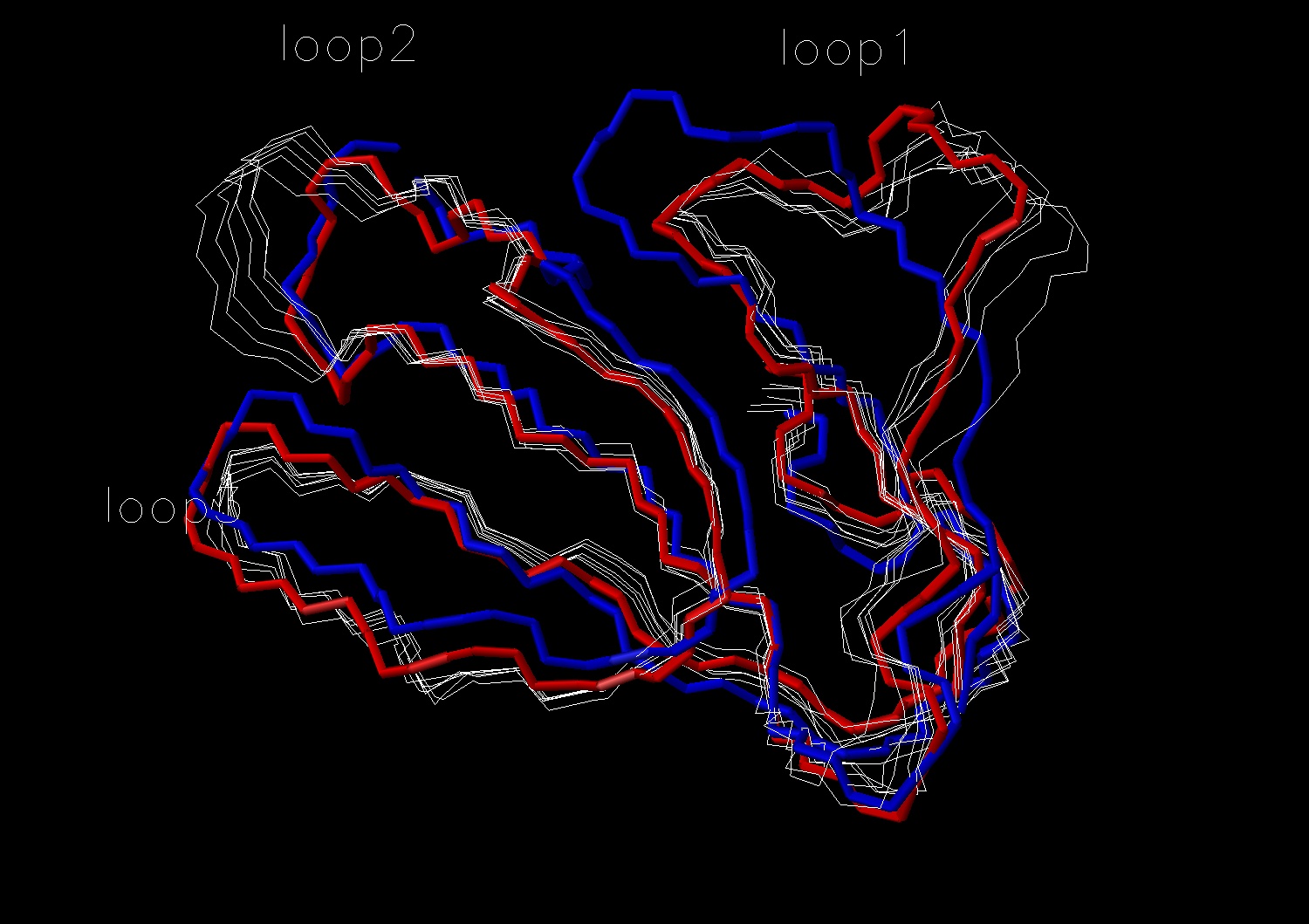
Fig. 6 Fit of the structures with highest RMSD (>2.0 A)
with fasciculin I (in red). For comparison fasciculin II (in blue) is also
shown. The conformational changes observed in loop I do not ressemble
those oberved between fasciculin I and fasciculin II.

Fig. 7 Fit of the structures with lowest RMSD (2.0
A) with fasciculin II (in blue). For comparison fasciculin I (in red) is
also
shown. The conformational changes observed in loop I do not ressemble
those oberved between fasciculin I and fasciculin II.
4.2 Fasciculin II (pdb 1fsc)4.1.2 ED analysis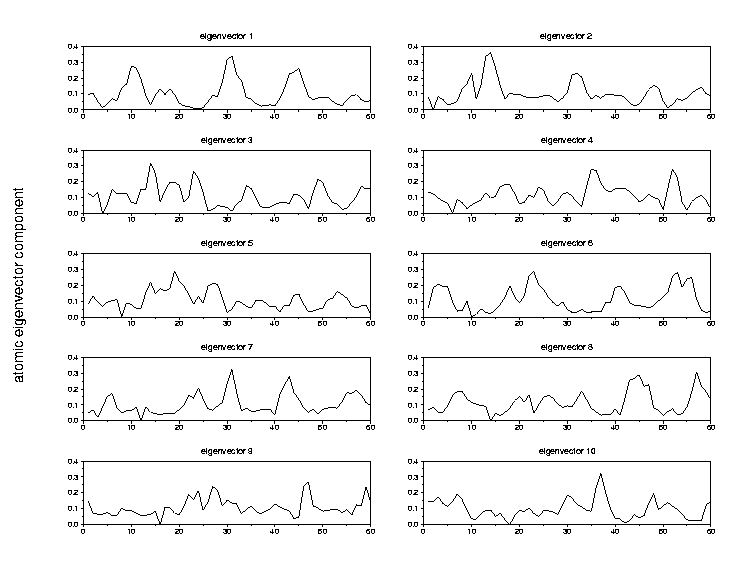
4.2.2. ED analysis