Tutorial: Using GRID for
Structure-based Drug Design
Rebecca Wade
European Media Laboratory,
Schloss-Wolfsbrunnenweg
33,
69118
Heidelberg,
Germany
rebecca.wade@eml.villa-bosch.de
22-05-2002
1
Introduction
GRID is one of the “original” programs for computational structure-based drug design. It provides a method to detect energetically favourable binding sites for functional groups on molecules of known structure. A chosen chemical group, a “probe”, is moved through a grid of points that is superimposed on the target molecule. At every position of the probe on the grid, its interaction energy with the target molecule is computed using empirical energy function. This interaction energy can be analysed by displaying energy contours using a molecular visualization program. GRID can be applied in a variety of ways including the following:
-detecting binding sites on macromolecules that can be used for the design of new ligands
-detecting sites that give selectivity between two macromolecules or ligands
-detecting hydration sites and designing to exclude/include interfacial water molecules
-docking molecules into macromolecular binding sites
-to compute molecular interaction fields for use in QSAR studies (on small molecules or macromolecules), including ADME predictions.
This practical consists of two parts.
-The first (greater01) is distributed with version 20 of the GRID program. It serves as a practical introduction into how to run GRID. This is optional.
-The second tutorial illustrates a real drug design example. GRID calculations will be performed that are similar to those that were the basis for the design of the anti-influenza drug Relenza® from Glaxo.
GRID version 20 will be used for the tutorial. This is available from http://www.moldiscovery.com/index.html. This version of GRID can be run on Linux and SGI workstations.
2
Tutorial 1: Greater01 – This tutorial
is Optional.
Follow the instructions at http://www.moldiscovery.com/manual/tut_greater01.html or locally at file /home/client1/programs/grid/manual/tut_greater01.html. Use the files in /home/client1/programs/grid/tutorials/greater01/.
Before starting, set up your environment by typing:
% source /home/client1/programs/grid/env_grid.sh
After starting Greater, check (in the File->Set Options pull-down menu) that the correct paths and directories are specified: GRID path: /home/client1/programs/grid/; grub.dat file: /home/client1/programs/grid/grub.dat.
Note that you should read in the file PDB.pdb with the PDB filter checkbox checked (not unchecked as stated in the tutorial).
3
Tutorial 2: Probing targets for anti-influenza agents
with GRID
The two targets studied here are the influenza neuraminidase (NA) and the influenza haemagglutinin (HA) proteins. GRID energy maps will be computed for an amino probe interacting with each of these targets. Discussion of these energy maps and the background to the structure-based design of anti-influenza agents can be found in reference 8 (pdf).
3.1
Neuraminidase
3.1.1
Examine
and prepare target coordinates
First make a directory (e.g. flu) for this tutorial and change into it.
% mkdir flu
% cd flu
We will take as the target the structure of neuraminidase determined by crystallography in complex with sialic acid (PDB identifier: 2bat). Take a look at it:
% cp
/home/client1/data/flu/2bat.brk .
% nedit 2bat.brk
&
The file 2bat.brk contains:
-Protein: Neuraminidase N2, from residue 82 to 469, 388 residues in total.
-Ligand: Sialic acid, SIA, residue 600.
-One Ca2+ ion: CA, HETATM residue 237
-Sugar molecules: NAG, MAN, FUC, NGL.
-Bound water molecules.
For comparative purposes, I have transformed the coordinates of neuraminidase from the PDB file 2bat so that the sialic acid in the binding site is superimposed upon the sialyl moiety in the structure of haemagglutinin in complex with 2-a-2,3-sialyllactose (1hgg1). This structure is in file 2batsup.pdb. Copy it to your current directory. It can be used by GREATER.
% cp
/home/client1/data/flu/2batsup.pdb .
% nedit 2batsup.pdb
&
3.1.2
Use
GREATER to add GRID parameters to target coordinates
Start GREATER:
% Greater
Click on the “Add single” icon.
In the pop-up window, enter the filename 2batsup.pdb, make sure the “PDB filter” box is ticked, and then click the OK button. (The “PDB filter” means that only “ATOM” records and not “HETATM” records will be used for the GRID calculation).
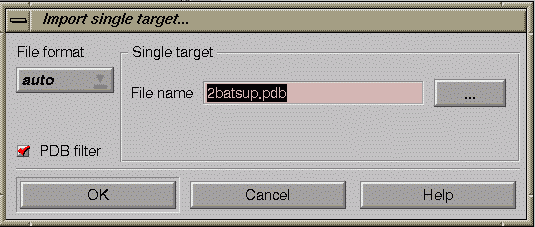
GREATER will run program GRIN to assign parameters to the atoms in the coordinate file and generate GRINLOUT and GRINKOUT. When “ready” is seen in the “status” column, view these files by right clicking on the 2batsup line and selecting “view text files”.
3.1.3
Use
GREATER to run GRID
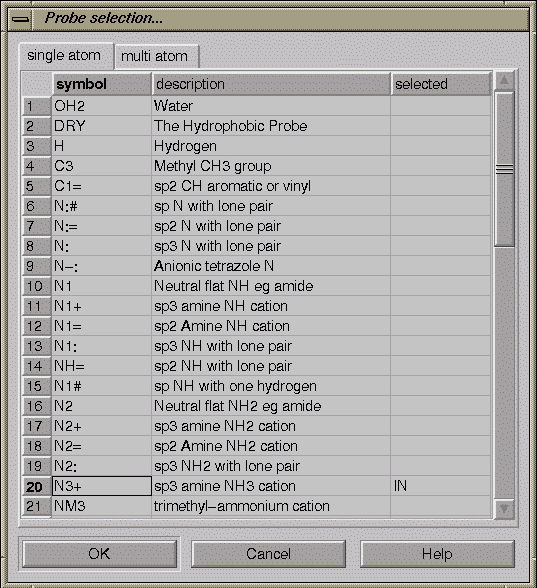
Click on the “Probes” icon to select a probe.
In the pop-up window, click on N3+ (row 20), and then click the OK button.
Set the parameters for the GRID run in the lower right-hand box:
-click “box”, then click NPLA and set to 2.0
-click “output”, then click LENG and set to 12 and click LIST and set to –2.
Define box size by left-clicking on the “Method” menu and selecting “Define box size”.
Untick the “automatic” box.
Set the box dimensions to the following values:
BOTX=-15.0, TOPX=3.0, BOTY=65.5,. TOPY=83.5, BOTZ=17.0, TOPZ=36.0
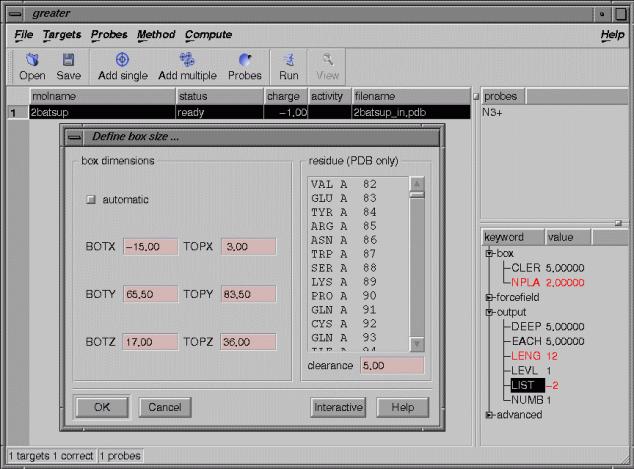
Click on the “Run” icon to run GRID.
In the pop-up window, enter “amino probe with neuraminidase” as the “Job title”, and click the “Run” button. This will take a few minutes to run interactively. A pop-up window will inform you when the job has finished and “completed” will appear in the “status” column.
3.1.4
Use
GREATER to view the results
Examine the text file output as before. The GRIDLOUT file gives selected information about the computed energies, including the location of the most energetically favourable grid point for the probe.
Then view the GRID map by right-clicking on the batsup line and selecting “view fields”. This will open a GVIEW window showing the protein and a grid box with contours.
-Add the coordinates of sialic acid (and other HETATMs) by using “Molecules Manager” in the Molecules pulldown menu. Click on “Add”, select “2batsup_out.pdb” and “Quit”.
-Alter the contour levels by using “Data Levels” in the “Data” pulldown menu. To compare with Figure 4a in reference 8, untick the positive values and set the negative value to –15 or –9 kcal/mol.
Can you see the favourable energy contour showing where the guanidine group of GG167 (Relenza) binds?
3.2
Haemagglutinin – This section is Optional
3.2.1
Examine
and prepare target coordinates
We will take as the target the structure of haemagglutinin determined by crystallography in complex with 2-a-2,3-sialyllactose (1hgg). Take a look at it:
% cp
/home/client1/data/flu/1hgg.brk .
% nedit 1hgg.brk
&
The file 1hgg.brk contains:
-Protein: Haemagglutin trimer with each monomer consisting of two chains, HA1 (328 residues) and HA2 (220 residues) (Subunits A-F).
-Ligand: Sialyllactose (NAN,GAL,GLC) in primary and secondary binding sites (Subunits G, H, I)
-N-linked carbohydrate: NAG, MAN
-Bound water molecules.
The primary binding site is not near the trimer interface so we will run GRID with a target that is a monomer rather than a trimer (just as we took a NA monomer as the target rather than an NA tetramer). The target will consist of subunits A, B and G corresponding to the first monomer in 1hgg.brk and its associated ligands, carbohydrates and water molecules.
1hgg.brk contains polar hydrogen atoms. The GRID energy function is best used without hydrogen coordinates entered for proteins so the H atom records have been made into remarks.
% sed ‘s/ATOM.........H/REMARK.......H/g’
< x.pdb > 1hgg1.pdb
The structure in file 1hgg1.pdb can be used by GREATER. Copy it to your current directory.
% cp
/home/client1/data/flu/1hgg1.pdb .
% nedit 1hgg1.pdb
&
3.2.2
Use
GREATER to add GRID parameters to target coordinates
Start GREATER again.
Click on the “Add single” icon. In the pop-up window, enter the filename 1hgg1.pdb, make sure the “PDB filter” box is ticked, and then click the OK button.
When GRIN has run, check the output text files.
3.2.3
Use
GREATER to run GRID
Do a run with the amino probe in exactly the same way as for NA using the same grid dimensions.
3.2.4
Use
GREATER to view the results
Examine the text file output as before. The GRIDLOUT file gives selected information about the computed energies, including the location of the most energetically favourable grid point for the probe.
How does the most favourable interaction energy of the amino probe in HA compare with that in NA?
Then view the GRID map by right-clicking on the 1hgg1 line and selecting “view fields”. This will open a GVIEW window showing the protein and a grid box with contours.
-Add the coordinates of sialic acid (and other HETATMs) by using “Molecules Manager” in the Molecules pulldown menu. Click on “Add”, select “1hgg1_out.pdb” and “Quit”.
-Alter the contour levels by using “Data Levels” in the “Data” pulldown menu. To compare with Figure 4a in reference 8, untick the positive values and set the negative value to–9 kcal/mol.
Can you see the favourable energy contour showing where
the amino group of Neu5Aca2Me9N
binds?
Compare this with the location of the ligand in 1hgj.brk for which the first monomer coordinates can be found in 1hgj1.pdb.
4 References
GRID Methodology:
- Goodford, P.J (1985) A Computational Procedure for Determining Energetically Favorable Binding Sites on Biologically Important Macromolecules. J. Med. Chem. (1985) 28, 849-857.
- Boobbyer,
D.N.A., Goodford, P.J., McWhinnie, P.M. and Wade, R.C. New Hydrogen-bond
Potentials for Use in Determining Energetically Favourable Binding Sites
on Molecules of Known Structure. J. Med. Chem. (1989) 32 1083-1094.
- Wade,R.C., Clark,K. and Goodford,P.J. Further Development of Hydrogen-Bond Functions for Use in Determining Energetically Favorable Binding Sites on Molecules of Known Structure. 1. Ligand Probe Groups with the Ability To Form Two Hydrogen Bonds J. Med. Chem. (1993) 36, 140-147.
- Wade,R.C.
and Goodford,P.J. Further
Development of Hydrogen-Bond Functions for Use in Determining
Energetically Favorable Binding Sites on Molecules of Known Structure. 2.
Ligand Probe Groups with the Ability To Form More Than Two Hydrogen
Bonds J. Med. Chem. (1993) 36, 148-156.
- Wade,
R.C. Molecular Interaction Fields.
In ``3D QSAR in Drug Design. Theory, Methods and Applications'' Ed.
Kubinyi,H. ESCOM, Leiden,
(1993), pp486-505.
- http://www.moldiscovery.com - GRID distribution and documentation.
Application to design of anti-influenza agents:
- von Itzstein, M., Wu, W.Y., Kok, G.B., Pegg, M.S., Dyason, J.C., Jin B, Van Phan, T., Smythe, M.L., White, H.F., Oliver, S.W., et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature (1993) 363, 418-23.(Medline)
Wade, R.C. 'Flu' and Structure Based Drug Design Structure (1997) 5, 1139-1146. (pdf)