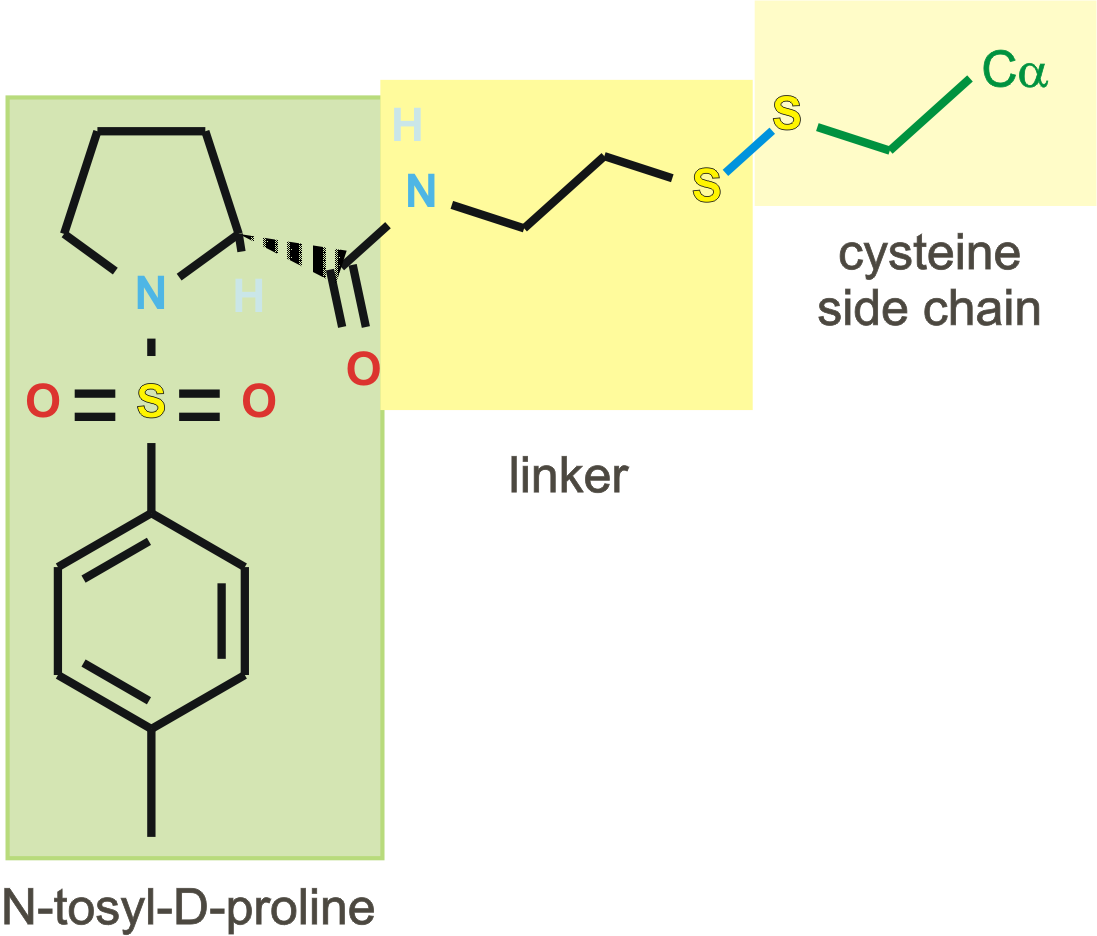
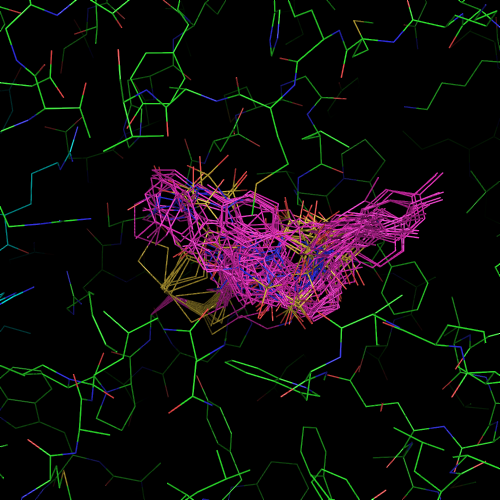
in silico tethering 2
(human TS)
download 1hvy.pdb
remove chain C and D, remove all HETATM - 1hvy-AB.pdb
load in pymol and addH - 1hvy-AB_addH.pdb
protonation with whatif (whatif.sh
1hvy-AB ) - 1hvy-AB_addH.wi.pdb
load 1hvy-AB_addH.wi.pdb in pymol and save again - 1hvy-AB_addH.wi.pdb (this step is important otherwise GOLD has problems with
atom types)
load 1hvy-AB_addH.wi.pdb in pymol and remove side chain of C195
in monomer A - 1hvy-AB-C195G.wi.pdb
center Ca-C195: 2710(too small!) - 25 A 1hvy-AB-C195G.wi.pdb :
2710CID_445503-cys3D.sdf :
43
start GOLD 3.1.1: gold_di1hvyC195G_wi.conf
WARNING: problems with atom types in mol2 for CME
/home/henricsn/LIGHTS/results/docking3_C195/ GoldScore
# res RMSD Fit iVDW iTOR eVDW iHB eHB covENE ENE iVDWw eVDWw iHBw eHBw covENEw
1 xray 4.504 -59.4402 3.1526 -5.7163 -40.1691 0.0000 0.0000 -1.6439 3.1526 -5.7163 -55.2326 0.0000 0.0000 -1.6439
2 xray 4.443 -59.8534 3.9711 -5.9469 -40.7504 0.0000 0.0000 -1.8458 3.9711 -5.9469 -56.0318 0.0000 0.0000 -1.8458
3 xray 5.462 -60.3346 4.1954 -5.5555 -42.0285 0.0000 0.4309 -1.6162 4.1954 -5.5555 -57.7892 0.0000 0.4309 -1.6162
4 xray 3.352 -60.6810 1.5995 -5.4443 -39.5667 0.0000 0.0000 -2.4320 1.5995 -5.4443 -54.4042 0.0000 0.0000 -2.4320
5 xray 3.645 -61.5485 2.4950 -6.1892 -40.9027 0.0000 0.0000 -1.6132 2.4950 -6.1892 -56.2412 0.0000 0.0000 -1.6132
6 xray 3.669 -61.7186 2.2291 -6.8185 -40.3690 0.0000 0.0300 -1.6518 2.2291 -6.8185 -55.5074 0.0000 0.0300 -1.6518
7 xray 5.535 -62.0732 3.8056 -5.6817 -42.7412 0.0000 0.0000 -1.4280 3.8056 -5.6817 -58.7691 0.0000 0.0000 -1.4280
8 xray 2.822 -62.1939 2.1269 -6.3893 -40.9256 0.0000 0.0000 -1.6588 2.1269 -6.3893 -56.2727 0.0000 0.0000 -1.6588
9 xray 2.568 -62.4729 2.2320 -5.8512 -41.4408 0.0000 0.3061 -2.1788 2.2320 -5.8512 -56.9810 0.0000 0.3061 -2.1788
10 xray 6.318 -62.5752 4.1789 -6.1731 -43.5092 0.0000 1.1532 -1.9090 4.1789 -6.1731 -59.8252 0.0000 1.1532 -1.9090
11 xray 2.387 -62.6077 1.5828 -6.4519 -40.4765 0.0000 0.0000 -2.0834 1.5828 -6.4519 -55.6551 0.0000 0.0000 -2.0834
12 xray 4.966 -62.6177 2.3765 -8.7283 -39.7791 0.0000 0.0000 -1.5697 2.3765 -8.7283 -54.6962 0.0000 0.0000 -1.5697
13 xray 2.616 -62.6683 2.3732 -6.0192 -41.5939 0.0000 0.3136 -2.1442 2.3732 -6.0192 -57.1916 0.0000 0.3136 -2.1442
14 xray 3.340 -62.9468 1.9027 -7.0264 -40.8758 0.0000 0.0000 -1.6189 1.9027 -7.0264 -56.2042 0.0000 0.0000 -1.6189
15 xray 3.975 -62.9637 1.4264 -7.0331 -40.6843 0.0000 0.0000 -1.4161 1.4264 -7.0331 -55.9409 0.0000 0.0000 -1.4161
16 xray 5.644 -62.9757 5.5534 -6.0329 -44.1881 0.0000 0.0000 -1.7376 5.5534 -6.0329 -60.7586 0.0000 0.0000 -1.7376
17 xray 2.972 -63.0394 2.2253 -5.6718 -41.5101 0.0000 0.0000 -2.5166 2.2253 -5.6718 -57.0764 0.0000 0.0000 -2.5166
18 xray 4.595 -63.5407 2.0683 -6.8584 -41.0213 0.0000 0.0000 -2.3464 2.0683 -6.8584 -56.4042 0.0000 0.0000 -2.3464
19 xray 6.261 -64.2663 3.7663 -7.5552 -42.4536 0.0000 0.0000 -2.1036 3.7663 -7.5552 -58.3738 0.0000 0.0000 -2.1036
20 xray 3.384 -64.4068 1.6544 -7.2204 -41.4309 0.0000 0.0000 -1.8733 1.6544 -7.2204 -56.9675 0.0000 0.0000 -1.8733
21 xray 6.469 -64.4246 2.3275 -7.6638 -42.5443 0.0000 1.3282 -1.9182 2.3275 -7.6638 -58.4984 0.0000 1.3282 -1.9182
22 xray 3.772 -64.4524 1.7363 -7.7362 -40.0946 0.0000 0.0000 -3.3225 1.7363 -7.7362 -55.1301 0.0000 0.0000 -3.3225
23 xray 2.632 -64.9394 1.8613 -7.8494 -41.8114 0.0000 0.0000 -1.4607 1.8613 -7.8494 -57.4906 0.0000 0.0000 -1.4607
24 xray 3.177 -65.0240 2.2232 -6.8267 -41.6967 0.0000 0.0000 -3.0875 2.2232 -6.8267 -57.3330 0.0000 0.0000 -3.0875
25 xray 4.137 -65.0623 4.3791 -5.9340 -44.8109 0.0000 0.0000 -1.8925 4.3791 -5.9340 -61.6150 0.0000 0.0000 -1.8925
26 xray 2.855 -65.0641 2.5208 -6.3762 -43.2871 0.0000 0.0000 -1.6890 2.5208 -6.3762 -59.5197 0.0000 0.0000 -1.6890
27 xray 3.338 -65.1105 2.7025 -6.6793 -43.4392 0.0000 0.0000 -1.4047 2.7025 -6.6793 -59.7290 0.0000 0.0000 -1.4047
28 xray 2.481 -65.1859 1.7306 -9.8243 -40.4235 0.0000 0.0169 -1.5268 1.7306 -9.8243 -55.5823 0.0000 0.0169 -1.5268
29 xray 4.446 -65.2973 4.4042 -6.3207 -44.9447 0.0000 0.0000 -1.5818 4.4042 -6.3207 -61.7989 0.0000 0.0000 -1.5818
30 xray 3.458 -65.4956 2.8770 -5.5472 -44.5606 0.0000 0.0000 -1.5546 2.8770 -5.5472 -61.2708 0.0000 0.0000 -1.5546
31 xray 5.725 -65.6843 2.9628 -7.0739 -43.4977 0.0000 0.0733 -1.8371 2.9628 -7.0739 -59.8093 0.0000 0.0733 -1.8371
32 xray 4.845 -65.8752 4.4003 -7.2312 -44.2095 0.0000 0.0000 -2.2562 4.4003 -7.2312 -60.7881 0.0000 0.0000 -2.2562
33 xray 7.183 -66.0362 1.8648 -8.1044 -41.9527 0.0000 0.0000 -2.1116 1.8648 -8.1044 -57.6849 0.0000 0.0000 -2.1116
34 xray 3.986 -66.1226 1.5639 -7.6263 -42.3179 0.0000 0.0000 -1.8730 1.5639 -7.6263 -58.1870 0.0000 0.0000 -1.8730
35 xray 2.669 -66.4557 0.7338 -7.4513 -41.9312 0.0000 0.0000 -2.0827 0.7338 -7.4513 -57.6554 0.0000 0.0000 -2.0827
36 xray 3.383 -66.9269 0.5403 -6.4468 -42.3837 0.0000 0.0000 -2.7428 0.5403 -6.4468 -58.2776 0.0000 0.0000 -2.7428
37 xray 2.728 -66.9789 2.0932 -6.3552 -43.2045 0.0000 0.0000 -3.3108 2.0932 -6.3552 -59.4062 0.0000 0.0000 -3.3108
38 xray 5.225 -67.0699 1.5506 -8.6132 -42.7299 0.0000 0.2714 -1.5252 1.5506 -8.6132 -58.7536 0.0000 0.2714 -1.5252
39 xray 6.426 -67.2082 1.0637 -7.0772 -42.9409 0.0000 0.0000 -2.1509 1.0637 -7.0772 -59.0438 0.0000 0.0000 -2.1509
40 xray 3.768 -67.3073 -0.7698 -9.9821 -39.7107 0.0000 0.0000 -1.9532 -0.7698 -9.9821 -54.6022 0.0000 0.0000 -1.9532
41 xray 3.421 -67.4788 2.6678 -7.2102 -42.6710 0.0000 0.0000 -4.2638 2.6678 -7.2102 -58.6726 0.0000 0.0000 -4.2638
42 xray 5.386 -67.5075 -1.2621 -8.7764 -44.2361 0.0000 5.3057 -1.9501 -1.2621 -8.7764 -60.8246 0.0000 5.3057 -1.9501
43 xray 3.235 -67.7542 2.8209 -5.6635 -45.7922 0.0000 0.0000 -1.9474 2.8209 -5.6635 -62.9642 0.0000 0.0000 -1.9474
44 xray 2.908 -67.8293 1.2771 -10.4678 -41.6203 0.0000 0.1099 -1.5207 1.2771 -10.4678 -57.2279 0.0000 0.1099 -1.5207
45 xray 2.695 -67.9202 0.1980 -6.7943 -42.6999 0.0000 0.0000 -2.6116 0.1980 -6.7943 -58.7123 0.0000 0.0000 -2.6116
46 xray 4.892 -68.0959 1.9908 -11.6670 -41.1389 0.0000 0.0000 -1.8537 1.9908 -11.6670 -56.5659 0.0000 0.0000 -1.8537
47 xray 2.697 -68.2822 1.7318 -10.5038 -41.4919 0.0000 0.0000 -2.4590 1.7318 -10.5038 -57.0513 0.0000 0.0000 -2.4590
48 xray 2.836 -68.4982 0.8810 -6.8182 -44.7829 0.0000 0.9548 -1.9394 0.8810 -6.8182 -61.5765 0.0000 0.9548 -1.9394
49 xray 5.884 -71.3459 2.0793 -9.7323 -45.1209 0.0000 0.0000 -1.6516 2.0793 -9.7323 -62.0413 0.0000 0.0000 -1.6516
50 xray 2.695 -73.1798 1.6583 -11.1910 -44.7009 0.0000 0.3930 -2.5765 1.6583 -11.1910 -61.4637 0.0000 0.3930 -2.5765
GOLD CONFIGURATION FILE generated by gold front
end (GOLD v3.1.1) POPULATION popsiz = 100 select_pressure = 1.1 n_islands = 5 maxops = 100000 niche_siz = 2 GENETIC OPERATORS pt_crosswt = 95 allele_mutatewt = 95 migratewt = 10 FLOOD FILL radius = 15 origin = 4.869 -6.842 7.681 do_cavity = 1 floodfill_atom_no = 2710 cavity_file = bmode.mol2 floodfill_center = atom DATA FILES protein_datafile =
1hvy-AB-C195G_wi.pdb ligand_data_file
CID_445503-cys3D.sdf 50 param_file = DEFAULT set_ligand_atom_types = 1 set_protein_atom_types = 1 directory = . tordist_file = DEFAULT make_subdirs = 0 save_lone_pairs = 1 fit_points_file = fit_pts.mol2 read_fitpts = 0 FLAGS display = 0 internal_ligand_h_bonds = 0 n_ligand_bumps = 0 flip_free_corners = 0 flip_amide_bonds = 0 flip_planar_n = 1 flip_ring_NRR
flip_ring_NHR flip_pyramidal_n = 0 rotate_carboxylic_oh = flip use_tordist = 1 TERMINATION early_termination = 0 n_top_solutions = 3 rms_tolerance = 1.5 CONSTRAINTS force_constraints = 0 COVALENT BONDING covalent = 1 covalent_protein_atom_no = 2710 covalent_ligand_atom_no = 43 SAVE OPTIONS save_score_in_file = 1 save_protein_torsions = 1 concatenated_output =
/home/henricsn/LIGHTS/results/docking3_C195/docking_di1hvyC195G_wi_445503.sdf clean_up_option
delete_all_solutions clean_up_option
delete_redundant_log_files clean_up_option
delete_empty_directories output_file_format = MACCS FITNESS FUNCTION SETTINGS initial_virtual_pt_match_max = 2.5 relative_ligand_energy = 0 score_param_file = DEFAULT start_vdw_linear_cutoff = 4 Privacy Imprint