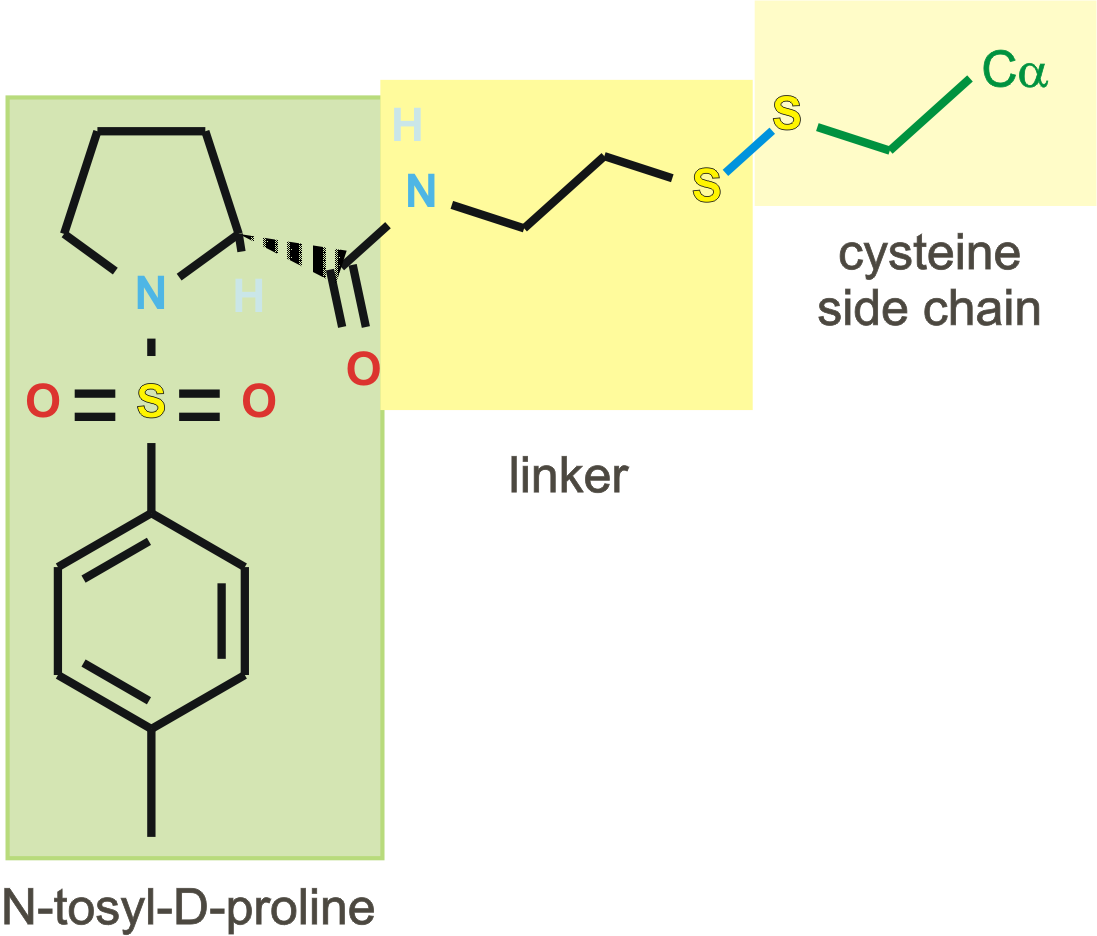
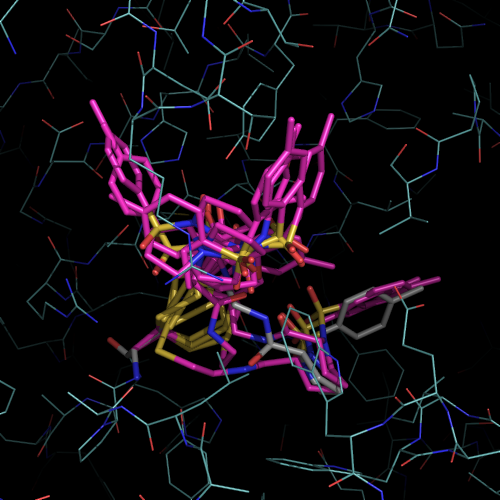
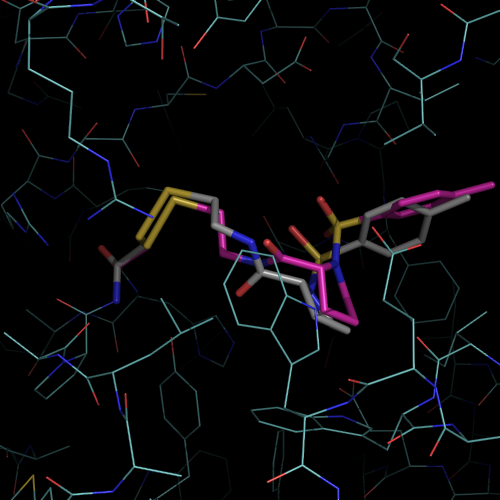
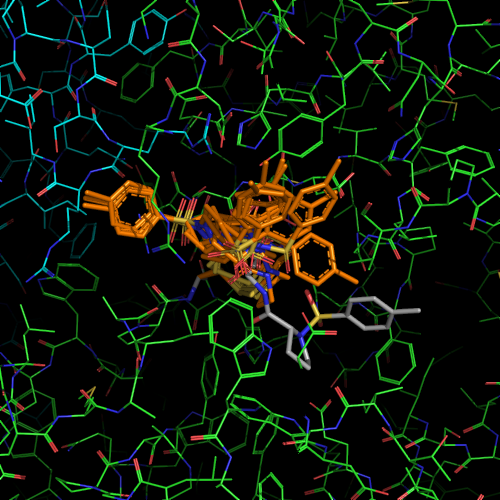
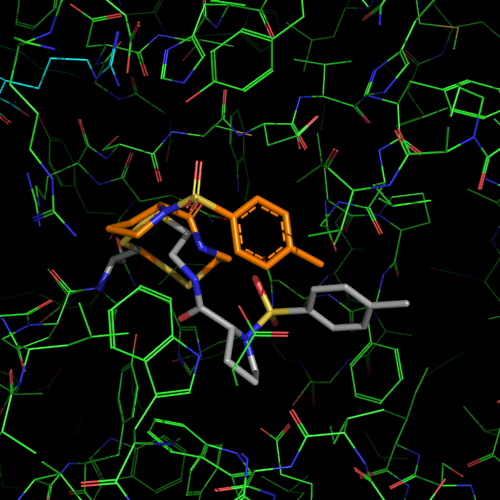
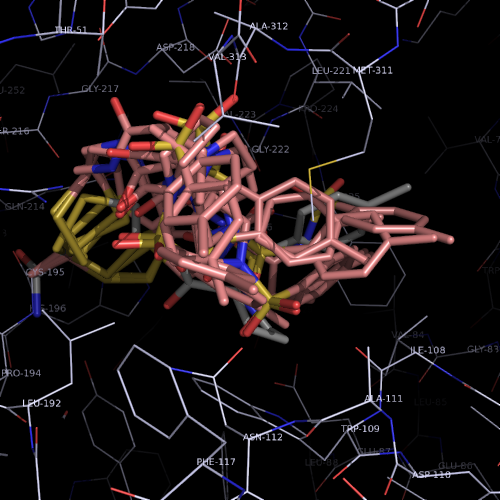
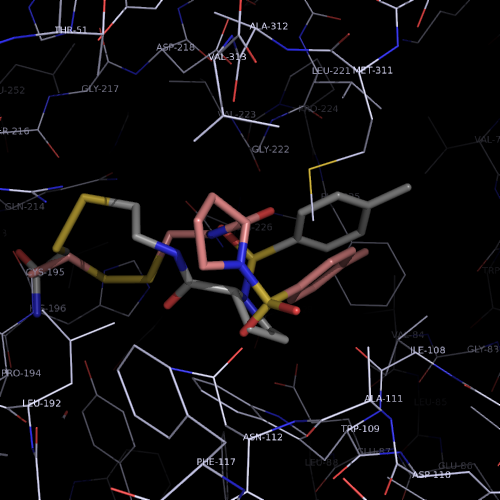
| PDB |
Name |
SG |
Res |
monomers /AU |
|
| 1f4b | native |
I213 |
1.75 |
1 |
without ligand |
| 1f4c | C146 tethered N-tosyl-D-proline | P63 |
2.0 |
2 |
covalent bound to active site
cysteine C146 |
| 1f4d | L143C tethered N-tosyl-D-proline | P63 | 2.15 |
2 |
covalent bound to new cysteine
L143C, C146S |
| 1f4e | Noncovalent N-tosyl-D-proline |
I213 | 1.90 |
1 |
|
| 1f4f | Glu-TP |
P63 | 2.0 |
2 |
|
| 1f4g | Glu-TP-beta-Ala |
P63 | 1.75 |
2 |