docking 7
(E. coli TS)
Docking of N-tosyl-D-proline to dimer of E. coli TS (1f4f.pdb, chain A and B)
- download 1f4f.pdb
- superimpose to 1hvy-AB_addH.wi.pdb
(sup2pdbs_sh
1hvy-AB_addH.wi.pdb 1f4f.pdb)
- 1f4f.pdbf
- load in pymol, correct bonds of ligand and addH - 1f4f_addH.wat.pdb
- protonation with whatif using water from X-ray structure (whatif.sh
1f4d_addh.wat) - 1f4f_addh.wat.wi.pdb
- remove water molecules and sulfate - 1f4f_addh.wi.pdb
- load 1f4f_addh.wi.pdb in
pymol and save again
(this step is important otherwise GOLD has problems with
atom types)
- center Ca-C146: 2341
(center 1hvy-Ca-C195: 3.002
2.973 11.352)
radius: 25 A
- start GOLD 3.1.1: gold_di1f4f_wi.conf
ligand: CID_445505-3D.sdf
- WARNING: problems with atom types in mol2
output: docking_di1f4f_wi_445505.25A.sdf
ChemScore
gold_analysis.sh -xray1 1f4f_CID_445505.pdb -file docking_di1f4f_wi_445505.CS25A.sdf
# res RMSD Fit DG ZeCo HB MET LIPO iHB DEC DEI ROT COVA HBw METw LIPOw iHBw DECw DEIw ROTw
1 xray 1.753 27.4186 -31.3544 -5.4800 5.0946 0.0000 155.1282 0.0000 1.4337 2.5021 3.6295 undef -17.0159 -0.0000 -18.1500 0.0000 1.4337 2.5021 9.2915
2 xray 1.659 25.0847 -27.7759 -5.4800 4.1726 0.0000 150.8639 0.0000 0.0653 2.6259 3.6295 undef -13.9363 -0.0000 -17.6511 0.0000 0.0653 2.6259 9.2915
3 xray 1.896 25.0461 -29.0250 -5.4800 4.4406 0.0000 153.8873 0.0000 0.0361 3.9428 3.6295 undef -14.8317 -0.0000 -18.0048 0.0000 0.0361 3.9428 9.2915
4 xray 1.832 24.7055 -27.8444 -5.4800 4.4007 0.0000 144.9352 0.0000 0.1681 2.9707 3.6295 undef -14.6985 -0.0000 -16.9574 0.0000 0.1681 2.9707 9.2915
5 xray 1.576 24.6838 -28.0006 -5.4800 4.1925 0.0000 152.2144 0.0000 0.4910 2.8258 3.6295 undef -14.0030 -0.0000 -17.8091 0.0000 0.4910 2.8258 9.2915
6 xray 1.919 24.5754 -29.3954 -5.4800 4.8737 0.0000 144.6895 0.0000 0.4359 4.3841 3.6295 undef -16.2782 -0.0000 -16.9287 0.0000 0.4359 4.3841 9.2915
7 xray 2.069 24.5582 -27.7958 -5.4800 4.3099 0.0000 147.1144 0.0000 0.0427 3.1949 3.6295 undef -14.3949 -0.0000 -17.2124 0.0000 0.0427 3.1949 9.2915
8 xray 1.965 24.5008 -29.2215 -5.4800 4.8981 0.0000 142.5071 0.0000 0.1678 4.5528 3.6295 undef -16.3597 -0.0000 -16.6733 0.0000 0.1678 4.5528 9.2915
9 xray 1.860 24.3195 -29.1858 -5.4800 4.8517 0.0000 143.5268 0.0000 0.3197 4.5467 3.6295 undef -16.2047 -0.0000 -16.7926 0.0000 0.3197 4.5467 9.2915
10 xray 1.416 24.2414 -29.6533 -5.4800 4.6528 0.0000 153.1991 0.0000 1.8516 3.5603 3.6295 undef -15.5405 -0.0000 -17.9243 0.0000 1.8516 3.5603 9.2915
11 xray 1.669 24.0346 -27.4963 -5.4800 4.1821 0.0000 148.2028 0.0000 0.0862 3.3755 3.6295 undef -13.9681 -0.0000 -17.3397 0.0000 0.0862 3.3755 9.2915
12 xray 1.789 23.8994 -26.7788 -5.4800 3.9727 0.0000 148.0455 0.0000 0.0288 2.8506 3.6295 undef -13.2689 -0.0000 -17.3213 0.0000 0.0288 2.8506 9.2915
13 xray 1.956 23.7914 -28.4613 -5.4800 4.4989 0.0000 147.4059 0.0000 0.1578 4.5122 3.6295 undef -15.0263 -0.0000 -17.2465 0.0000 0.1578 4.5122 9.2915
14 xray 1.785 23.6634 -27.8269 -5.4800 4.2526 0.0000 149.0155 0.0000 0.0798 4.0836 3.6295 undef -14.2036 -0.0000 -17.4348 0.0000 0.0798 4.0836 9.2915
15 xray 1.814 23.5843 -30.1534 -5.4800 4.9276 0.0000 149.6304 0.0000 0.3260 6.2432 3.6295 undef -16.4582 -0.0000 -17.5068 0.0000 0.3260 6.2432 9.2915
16 xray 2.126 23.2604 -28.3285 -5.4800 4.4166 0.0000 148.6198 0.0000 0.1872 4.8809 3.6295 undef -14.7514 -0.0000 -17.3885 0.0000 0.1872 4.8809 9.2915
17 xray 1.947 23.1834 -28.2599 -5.4800 4.1017 0.0000 157.0222 0.0000 1.9202 3.1563 3.6295 undef -13.6998 -0.0000 -18.3716 0.0000 1.9202 3.1563 9.2915
18 xray 2.060 23.0144 -30.0224 -5.4800 4.6164 0.0000 157.3930 0.0000 3.2549 3.7531 3.6295 undef -15.4189 -0.0000 -18.4150 0.0000 3.2549 3.7531 9.2915
19 xray 2.289 22.9514 -27.3890 -5.4800 4.0851 0.0000 150.0542 0.0000 0.2907 4.1468 3.6295 undef -13.6441 -0.0000 -17.5563 0.0000 0.2907 4.1468 9.2915
20 xray 9.119 22.5538 -26.4070 -5.4800 4.8255 0.0000 120.5235 0.0000 0.7173 3.1359 3.6295 undef -16.1173 -0.0000 -14.1013 0.0000 0.7173 3.1359 9.2915
21 xray 2.961 22.5445 -28.9133 -5.4800 4.8073 0.0000 142.4636 0.0000 1.3114 5.0574 3.6295 undef -16.0565 -0.0000 -16.6682 0.0000 1.3114 5.0574 9.2915
22 xray 2.543 22.3236 -25.1957 -5.4800 3.7526 0.0000 140.7996 0.0000 0.0662 2.8058 3.6295 undef -12.5336 -0.0000 -16.4736 0.0000 0.0662 2.8058 9.2915
23 xray 2.408 22.1286 -26.0876 -5.4800 3.9160 0.0000 143.7578 0.0000 0.1219 3.8370 3.6295 undef -13.0794 -0.0000 -16.8197 0.0000 0.1219 3.8370 9.2915
24 xray 9.537 22.0551 -25.7978 -5.4800 4.4548 0.0000 125.8996 0.0000 0.0431 3.6996 3.6295 undef -14.8790 -0.0000 -14.7303 0.0000 0.0431 3.6996 9.2915
25 xray 1.930 21.8445 -27.9396 -5.4800 4.4413 0.0000 144.5900 0.0000 1.7003 4.3948 3.6295 undef -14.8341 -0.0000 -16.9170 0.0000 1.7003 4.3948 9.2915
26 xray 2.045 21.8066 -26.2197 -5.4800 3.8941 0.0000 145.5122 0.0000 0.0539 4.3592 3.6295 undef -13.0062 -0.0000 -17.0249 0.0000 0.0539 4.3592 9.2915
27 xray 2.033 21.5693 -26.3262 -5.4800 3.9602 0.0000 144.5355 0.0000 1.5603 3.1966 3.6295 undef -13.2270 -0.0000 -16.9106 0.0000 1.5603 3.1966 9.2915
28 xray 2.429 20.7520 -25.3458 -5.4800 3.9881 0.0000 135.3584 0.0000 0.9598 3.6339 3.6295 undef -13.3203 -0.0000 -15.8369 0.0000 0.9598 3.6339 9.2915
29 xray 2.853 18.8275 -21.5400 -5.4800 2.5437 0.0000 144.0645 0.0000 0.0209 2.6917 3.6295 undef -8.4960 -0.0000 -16.8555 0.0000 0.0209 2.6917 9.2915
30 xray 2.913 18.5990 -22.7024 -5.4800 2.9796 0.0000 141.5563 0.0000 0.4726 3.6308 3.6295 undef -9.9518 -0.0000 -16.5621 0.0000 0.4726 3.6308 9.2915
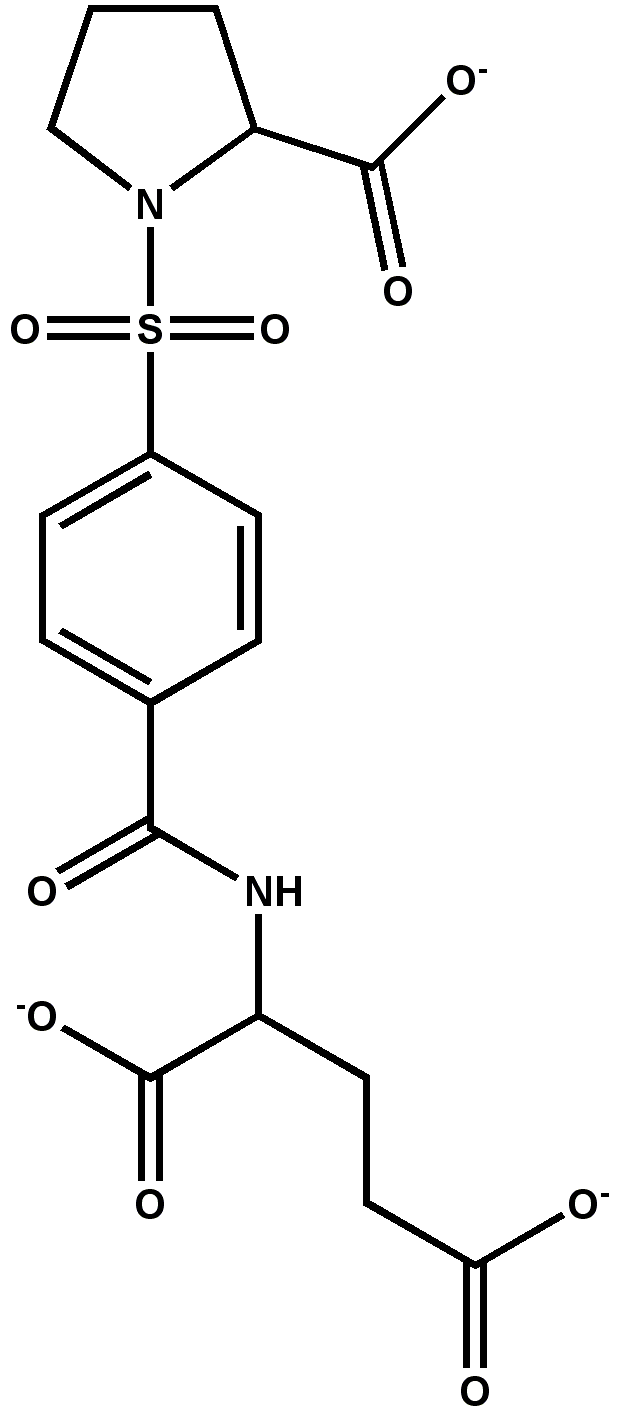
Docking of N-tosyl-D-proline to dimer of E. coli TS (1f4g.pdb, chain A and B)
- download 1f4f.pdb
- superimpose to 1hvy-AB_addH.wi.pdb
(sup2pdbs_sh
1hvy-AB_addH.wi.pdb 1f4f.pdb)
- 1f4g.pdbf
- load in pymol, correct bonds of ligand and addH - 1f4g_addH.wat.pdb
- protonation with whatif using water from X-ray structure (whatif.sh
1f4d_addh.wat) - 1f4g_addh.wat.wi.pdb
- remove water molecules and sulfate - 1f4g_addh.wi.pdb
- load 1f4g_addh.wi.pdb in
pymol and save again
(this step is important otherwise GOLD has problems with
atom types)
- center Ca-C146: 2317
(center 1hvy-Ca-C195: 3.002
2.973 11.352)
radius: 25 A
- start GOLD 3.1.1: gold_di1f4f_wi.conf
ligand: CID_445506-3D.sdf
- WARNING: problems with atom types in mol2
output: docking_di1f4g_wi_445506.25A.sdf
/home/henricsn/LIGHTS/results/docking3_C195/
ChemScore
The ligand f4g_CID_445506.pdb has two alternative conformations!
gold_analysis.sh -xray1 1f4g_A-CID_445506.pdb -file docking_di1f4g_wi_445506.CS25A.sdf
# res RMSD Fit DG ZeCo HB MET LIPO iHB DEC DEI ROT COVA HBw METw LIPOw iHBw DECw DEIw ROTw
1 xray 1.971 27.4159 -32.1422 -5.4800 5.7733 0.0000 170.7340 0.0000 0.2483 4.4779 4.9206 undef -19.2829 -0.0000 -19.9759 0.0000 0.2483 4.4779 12.5966
2 xray 1.789 27.0564 -31.3946 -5.4800 5.3023 0.0000 177.7917 0.0000 0.3287 4.0095 4.9206 undef -17.7096 -0.0000 -20.8016 0.0000 0.3287 4.0095 12.5966
3 xray 2.734 27.0033 -30.0221 -5.4800 5.8334 0.0000 150.8973 0.0000 0.0686 2.9502 4.9206 undef -19.4837 -0.0000 -17.6550 0.0000 0.0686 2.9502 12.5966
4 xray 1.897 26.6531 -29.8419 -5.4800 4.8257 0.0000 178.1256 0.0000 0.0218 3.1670 4.9206 undef -16.1178 -0.0000 -20.8407 0.0000 0.0218 3.1670 12.5966
5 xray 2.319 26.2226 -30.0742 -5.4800 4.9540 0.0000 176.4492 0.0000 0.2276 3.6240 4.9206 undef -16.5462 -0.0000 -20.6446 0.0000 0.2276 3.6240 12.5966
6 xray 1.769 25.2142 -30.6074 -5.4800 5.2451 0.0000 172.6942 0.0000 1.0626 4.3306 4.9206 undef -17.5188 -0.0000 -20.2052 0.0000 1.0626 4.3306 12.5966
7 xray 3.094 24.9121 -29.9464 -5.4800 5.4772 0.0000 160.4200 0.0000 1.2644 3.7699 4.9206 undef -18.2939 -0.0000 -18.7691 0.0000 1.2644 3.7699 12.5966
8 xray 2.106 24.5880 -32.0059 -5.4800 5.8059 0.0000 168.6404 0.0000 1.8352 5.5827 4.9206 undef -19.3916 -0.0000 -19.7309 0.0000 1.8352 5.5827 12.5966
9 xray 1.593 24.4671 -31.2821 -5.4800 5.5154 0.0000 170.7454 0.0000 1.1443 5.6707 4.9206 undef -18.4215 -0.0000 -19.9772 0.0000 1.1443 5.6707 12.5966
10 xray 2.665 24.4500 -28.0129 -5.4800 5.4301 0.0000 145.2392 0.0000 0.0004 3.5625 4.9206 undef -18.1366 -0.0000 -16.9930 0.0000 0.0004 3.5625 12.5966
11 xray 2.510 24.2936 -27.6299 -5.4800 4.9325 0.0000 156.1693 0.0000 0.0212 3.3151 4.9206 undef -16.4747 -0.0000 -18.2718 0.0000 0.0212 3.3151 12.5966
12 xray 2.017 24.0883 -27.8966 -5.4800 4.9321 0.0000 158.4617 0.0000 0.0883 3.7199 4.9206 undef -16.4732 -0.0000 -18.5400 0.0000 0.0883 3.7199 12.5966
13 xray 12.082 23.7730 -32.5613 -5.4800 5.9572 0.0000 169.0674 0.0000 3.1617 5.6267 4.9206 undef -19.8971 -0.0000 -19.7809 0.0000 3.1617 5.6267 12.5966
14 xray 9.941 23.6408 -27.5831 -5.4800 5.0946 0.0000 151.1423 0.0000 0.1263 3.8160 4.9206 undef -17.0160 -0.0000 -17.6836 0.0000 0.1263 3.8160 12.5966
15 xray 1.821 23.4942 -31.7764 -5.4800 5.4430 0.0000 177.0382 0.0000 0.9372 7.3450 4.9206 undef -18.1795 -0.0000 -20.7135 0.0000 0.9372 7.3450 12.5966
16 xray 2.330 23.1686 -26.3087 -5.4800 4.2907 0.0000 163.2000 0.0000 0.1310 3.0091 4.9206 undef -14.3309 -0.0000 -19.0944 0.0000 0.1310 3.0091 12.5966
17 xray 2.785 22.7572 -26.8896 -5.4800 4.9034 0.0000 150.6741 0.0000 0.2507 3.8817 4.9206 undef -16.3773 -0.0000 -17.6289 0.0000 0.2507 3.8817 12.5966
18 xray 2.343 22.6085 -28.7612 -5.4800 4.7872 0.0000 169.9872 0.0000 1.2080 4.9448 4.9206 undef -15.9893 -0.0000 -19.8885 0.0000 1.2080 4.9448 12.5966
19 xray 1.285 21.4518 -26.4631 -5.4800 3.7572 0.0000 179.7479 0.0000 0.3760 4.6353 4.9206 undef -12.5492 -0.0000 -21.0305 0.0000 0.3760 4.6353 12.5966
20 xray 2.712 21.2189 -26.4984 -5.4800 4.9593 0.0000 145.7356 0.0000 0.9886 4.2909 4.9206 undef -16.5640 -0.0000 -17.0511 0.0000 0.9886 4.2909 12.5966
21 xray 2.454 20.8927 -26.0712 -5.4800 3.9243 0.0000 171.6306 0.0000 0.1379 5.0406 4.9206 undef -13.1071 -0.0000 -20.0808 0.0000 0.1379 5.0406 12.5966
22 xray 2.868 20.8533 -24.5943 -5.4800 3.6754 0.0000 166.1109 0.0000 0.7125 3.0286 4.9206 undef -12.2760 -0.0000 -19.4350 0.0000 0.7125 3.0286 12.5966
23 xray 1.487 20.6526 -27.0064 -5.4800 4.1147 0.0000 174.1864 0.0000 0.8345 5.5193 4.9206 undef -13.7432 -0.0000 -20.3798 0.0000 0.8345 5.5193 12.5966
24 xray 2.956 19.9155 -23.8049 -5.4800 3.9812 0.0000 150.6340 0.0000 0.0634 3.8260 4.9206 undef -13.2974 -0.0000 -17.6242 0.0000 0.0634 3.8260 12.5966
25 xray 10.858 18.9303 -28.8118 -5.4800 5.0464 0.0000 163.0200 0.0000 4.3058 5.5757 4.9206 undef -16.8551 -0.0000 -19.0733 0.0000 4.3058 5.5757 12.5966
26 xray 2.486 18.4590 -23.3054 -5.4800 3.8391 0.0000 150.4229 0.0000 1.4824 3.3640 4.9206 undef -12.8225 -0.0000 -17.5995 0.0000 1.4824 3.3640 12.5966
27 xray 2.191 18.0896 -25.4309 -5.4800 3.6681 0.0000 173.4712 0.0000 3.2218 4.1195 4.9206 undef -12.2514 -0.0000 -20.2961 0.0000 3.2218 4.1195 12.5966
28 xray 10.881 16.8396 -24.9012 -5.4800 4.3556 0.0000 149.3180 0.0000 2.6052 5.4564 4.9206 undef -14.5476 -0.0000 -17.4702 0.0000 2.6052 5.4564 12.5966
29 xray 10.774 15.9560 -24.6126 -5.4800 4.2197 0.0000 150.7293 0.0000 4.0450 4.6116 4.9206 undef -14.0939 -0.0000 -17.6353 0.0000 4.0450 4.6116 12.5966
30 xray 11.721 13.9472 -18.9991 -5.4800 2.8623 0.0000 141.5021 0.0000 0.1042 4.9478 4.9206 undef -9.5600 -0.0000 -16.5558 0.0000 0.1042 4.9478 12.5966
gold_analysis.sh -xray1 1f4g_B-CID_445506.pdb -file docking_di1f4g_wi_445506.CS25A.sdf
# res RMSD Fit DG ZeCo HB MET LIPO iHB DEC DEI ROT COVA HBw METw LIPOw iHBw DECw DEIw ROTw
xray 1.886 27.4159 -32.1422 -5.4800 5.7733 0.0000 170.7340 0.0000 0.2483 4.4779 4.9206 undef -19.2829 -0.0000 -19.9759 0.0000 0.2483 4.4779 12.5966
2 xray 1.715 27.0564 -31.3946 -5.4800 5.3023 0.0000 177.7917 0.0000 0.3287 4.0095 4.9206 undef -17.7096 -0.0000 -20.8016 0.0000 0.3287 4.0095 12.5966
3 xray 2.651 27.0033 -30.0221 -5.4800 5.8334 0.0000 150.8973 0.0000 0.0686 2.9502 4.9206 undef -19.4837 -0.0000 -17.6550 0.0000 0.0686 2.9502 12.5966
4 xray 1.829 26.6531 -29.8419 -5.4800 4.8257 0.0000 178.1256 0.0000 0.0218 3.1670 4.9206 undef -16.1178 -0.0000 -20.8407 0.0000 0.0218 3.1670 12.5966
5 xray 2.278 26.2226 -30.0742 -5.4800 4.9540 0.0000 176.4492 0.0000 0.2276 3.6240 4.9206 undef -16.5462 -0.0000 -20.6446 0.0000 0.2276 3.6240 12.5966
6 xray 1.693 25.2142 -30.6074 -5.4800 5.2451 0.0000 172.6942 0.0000 1.0626 4.3306 4.9206 undef -17.5188 -0.0000 -20.2052 0.0000 1.0626 4.3306 12.5966
7 xray 3.074 24.9121 -29.9464 -5.4800 5.4772 0.0000 160.4200 0.0000 1.2644 3.7699 4.9206 undef -18.2939 -0.0000 -18.7691 0.0000 1.2644 3.7699 12.5966
8 xray 2.034 24.5880 -32.0059 -5.4800 5.8059 0.0000 168.6404 0.0000 1.8352 5.5827 4.9206 undef -19.3916 -0.0000 -19.7309 0.0000 1.8352 5.5827 12.5966
9 xray 1.435 24.4671 -31.2821 -5.4800 5.5154 0.0000 170.7454 0.0000 1.1443 5.6707 4.9206 undef -18.4215 -0.0000 -19.9772 0.0000 1.1443 5.6707 12.5966
10 xray 2.618 24.4500 -28.0129 -5.4800 5.4301 0.0000 145.2392 0.0000 0.0004 3.5625 4.9206 undef -18.1366 -0.0000 -16.9930 0.0000 0.0004 3.5625 12.5966
11 xray 2.429 24.2936 -27.6299 -5.4800 4.9325 0.0000 156.1693 0.0000 0.0212 3.3151 4.9206 undef -16.4747 -0.0000 -18.2718 0.0000 0.0212 3.3151 12.5966
12 xray 1.910 24.0883 -27.8966 -5.4800 4.9321 0.0000 158.4617 0.0000 0.0883 3.7199 4.9206 undef -16.4732 -0.0000 -18.5400 0.0000 0.0883 3.7199 12.5966
13 xray 11.628 23.7730 -32.5613 -5.4800 5.9572 0.0000 169.0674 0.0000 3.1617 5.6267 4.9206 undef -19.8971 -0.0000 -19.7809 0.0000 3.1617 5.6267 12.5966
14 xray 9.441 23.6408 -27.5831 -5.4800 5.0946 0.0000 151.1423 0.0000 0.1263 3.8160 4.9206 undef -17.0160 -0.0000 -17.6836 0.0000 0.1263 3.8160 12.5966
15 xray 1.721 23.4942 -31.7764 -5.4800 5.4430 0.0000 177.0382 0.0000 0.9372 7.3450 4.9206 undef -18.1795 -0.0000 -20.7135 0.0000 0.9372 7.3450 12.5966
16 xray 2.284 23.1686 -26.3087 -5.4800 4.2907 0.0000 163.2000 0.0000 0.1310 3.0091 4.9206 undef -14.3309 -0.0000 -19.0944 0.0000 0.1310 3.0091 12.5966
17 xray 2.704 22.7572 -26.8896 -5.4800 4.9034 0.0000 150.6741 0.0000 0.2507 3.8817 4.9206 undef -16.3773 -0.0000 -17.6289 0.0000 0.2507 3.8817 12.5966
18 xray 2.479 22.6085 -28.7612 -5.4800 4.7872 0.0000 169.9872 0.0000 1.2080 4.9448 4.9206 undef -15.9893 -0.0000 -19.8885 0.0000 1.2080 4.9448 12.5966
19 xray 1.343 21.4518 -26.4631 -5.4800 3.7572 0.0000 179.7479 0.0000 0.3760 4.6353 4.9206 undef -12.5492 -0.0000 -21.0305 0.0000 0.3760 4.6353 12.5966
20 xray 2.669 21.2189 -26.4984 -5.4800 4.9593 0.0000 145.7356 0.0000 0.9886 4.2909 4.9206 undef -16.5640 -0.0000 -17.0511 0.0000 0.9886 4.2909 12.5966
21 xray 2.400 20.8927 -26.0712 -5.4800 3.9243 0.0000 171.6306 0.0000 0.1379 5.0406 4.9206 undef -13.1071 -0.0000 -20.0808 0.0000 0.1379 5.0406 12.5966
22 xray 2.845 20.8533 -24.5943 -5.4800 3.6754 0.0000 166.1109 0.0000 0.7125 3.0286 4.9206 undef -12.2760 -0.0000 -19.4350 0.0000 0.7125 3.0286 12.5966
23 xray 1.409 20.6526 -27.0064 -5.4800 4.1147 0.0000 174.1864 0.0000 0.8345 5.5193 4.9206 undef -13.7432 -0.0000 -20.3798 0.0000 0.8345 5.5193 12.5966
24 xray 2.937 19.9155 -23.8049 -5.4800 3.9812 0.0000 150.6340 0.0000 0.0634 3.8260 4.9206 undef -13.2974 -0.0000 -17.6242 0.0000 0.0634 3.8260 12.5966
25 xray 10.431 18.9303 -28.8118 -5.4800 5.0464 0.0000 163.0200 0.0000 4.3058 5.5757 4.9206 undef -16.8551 -0.0000 -19.0733 0.0000 4.3058 5.5757 12.5966
26 xray 2.329 18.4590 -23.3054 -5.4800 3.8391 0.0000 150.4229 0.0000 1.4824 3.3640 4.9206 undef -12.8225 -0.0000 -17.5995 0.0000 1.4824 3.3640 12.5966
27 xray 2.148 18.0896 -25.4309 -5.4800 3.6681 0.0000 173.4712 0.0000 3.2218 4.1195 4.9206 undef -12.2514 -0.0000 -20.2961 0.0000 3.2218 4.1195 12.5966
28 xray 10.453 16.8396 -24.9012 -5.4800 4.3556 0.0000 149.3180 0.0000 2.6052 5.4564 4.9206 undef -14.5476 -0.0000 -17.4702 0.0000 2.6052 5.4564 12.5966
29 xray 10.338 15.9560 -24.6126 -5.4800 4.2197 0.0000 150.7293 0.0000 4.0450 4.6116 4.9206 undef -14.0939 -0.0000 -17.6353 0.0000 4.0450 4.6116 12.5966
30 xray 11.836 13.9472 -18.9991 -5.4800 2.8623 0.0000 141.5021 0.0000 0.1042 4.9478 4.9206 undef -9.5600 -0.0000 -16.5558 0.0000 0.1042 4.9478 12.5966
Privacy Imprint

